Mol:FL7AACGA0011
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
67 73 0 0 0 0 0 0 0 0999 V2000
-0.8281 2.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8281 1.3675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1134 0.9548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6013 1.3675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6013 2.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1134 2.6054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3160 0.9548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0307 1.3675 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0307 2.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3160 2.6054 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
2.7452 2.6052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4736 2.1846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2021 2.6052 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2021 3.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4736 3.8669 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7452 3.4464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5426 2.6052 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8079 3.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1134 0.1299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6904 0.7702 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9738 -0.3394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3224 0.1587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5916 -0.6095 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2664 -1.5063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9738 -1.9148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7495 -1.1295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0028 0.3343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2300 -1.4552 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6880 -1.6243 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6068 -2.4423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7033 0.6703 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2194 -0.0110 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9625 0.2781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6796 0.2858 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1585 0.8070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5083 0.4639 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6019 -0.0102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5547 -0.2113 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.1894 0.5875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4750 4.5001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2548 -2.9978 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1749 -3.0744 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3020 -3.7039 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9887 -3.4368 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6512 -3.4296 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1697 -2.9481 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5689 -3.2651 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7248 -3.2333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4524 -3.6901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5897 -3.8512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0174 -2.7063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7180 -2.7040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5648 -2.1426 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5648 -1.4187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2716 -2.6909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0665 -2.1874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7424 -2.6073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3952 -2.2303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0481 -2.6073 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0481 -3.3611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3952 -3.7381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7424 -3.3611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6159 -3.5532 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5470 -2.1436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.1894 -1.0310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3971 -4.4075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3413 -4.5001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
32 37 1 0 0 0 0
33 38 1 0 0 0 0
34 39 1 0 0 0 0
27 31 1 0 0 0 0
22 20 1 0 0 0 0
15 40 1 0 0 0 0
30 41 1 0 0 0 0
42 43 1 1 0 0 0
43 44 1 1 0 0 0
45 44 1 1 0 0 0
45 46 1 0 0 0 0
46 47 1 0 0 0 0
47 42 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 41 1 0 0 0 0
47 51 1 0 0 0 0
51 52 1 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
53 55 1 0 0 0 0
55 56 2 0 0 0 0
56 57 1 0 0 0 0
57 58 2 0 0 0 0
58 59 1 0 0 0 0
59 60 2 0 0 0 0
60 61 1 0 0 0 0
61 62 2 0 0 0 0
62 57 1 0 0 0 0
60 63 1 0 0 0 0
64 65 1 0 0 0 0
59 64 1 0 0 0 0
66 67 1 0 0 0 0
61 66 1 0 0 0 0
M CHG 1 10 1
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 64 65
M SBL 1 1 71
M SMT 1 ^ OCH3
M SBV 1 71 0.4989 -0.4636
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 66 67
M SBL 2 1 73
M SMT 2 OCH3
M SBV 2 73 0.0020 0.6694
S SKP 5
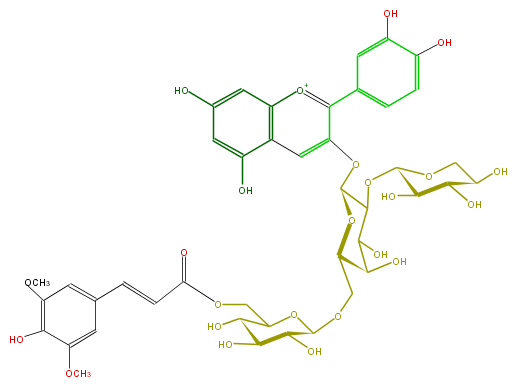
ID FL7AACGA0011
FORMULA C43H49O24
EXACTMASS 949.261377496
AVERAGEMASS 949.83476
SMILES Oc(c(OC)1)c(OC)cc(C=CC(OCC(C2O)OC(OCC(O3)C(O)C(O)C(OC(O7)C(O)C(O)C(C7)O)C3Oc(c4)c(c(c6)ccc(O)c6O)[o+1]c(c5)c4c(O)cc5O)C(C2O)O)=O)c1
M END