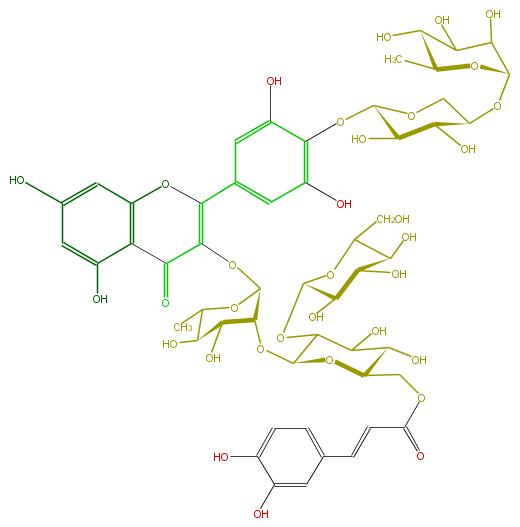
Mol:FL5FAGGS0017
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
86 94 0 0 0 0 0 0 0 0999 V2000
-4.4425 1.2987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4425 0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6838 -0.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9252 0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9252 1.2987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6838 1.7367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1665 -0.0153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4079 0.4226 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4079 1.2987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1665 1.7367 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1649 -0.8649 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6588 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1144 1.3146 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8877 1.7609 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8877 2.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1144 3.1003 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6588 2.6538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.3487 1.8220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6838 -0.7828 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6607 3.1001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7003 -0.0199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1144 3.9928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6607 1.3147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3650 3.4558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9128 2.7328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7017 3.0396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4630 3.0478 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9098 3.6011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2196 3.2368 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1427 2.7310 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3613 2.5063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0960 3.4428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3912 5.0917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7397 4.2353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5973 4.3293 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.3487 4.1366 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9502 4.8271 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1747 4.6506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6790 4.9584 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9352 4.4742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9289 5.4729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7993 5.3268 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3718 -1.0174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4931 -1.7046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9281 -1.2949 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2282 -1.2444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1068 -0.5570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6718 -0.9667 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2102 -2.0567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9904 -1.7544 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9317 -1.3849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0845 -1.8757 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1356 -1.5655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6083 -2.1940 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4243 -2.1089 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1668 -2.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6941 -1.8403 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8781 -1.9252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3498 -1.6342 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4188 -1.4641 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2940 -2.1010 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9294 -2.4358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8251 -0.4393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5460 -0.7835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0256 -0.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7451 0.1083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9581 0.5040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4456 -0.2433 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0329 -1.1783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8527 -0.2283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0803 0.5906 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4303 -2.9428 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0517 -3.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4066 -4.2135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2770 -3.5987 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9098 -4.2347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2646 -4.2348 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9069 -3.6151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1915 -3.6151 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1660 -4.2347 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1915 -4.8542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9069 -4.8542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8806 -4.2347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1657 -5.4729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5298 0.9874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8113 0.5210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
19 3 1 0 0 0 0
15 20 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
14 23 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
25 30 1 0 0 0 0
26 31 1 0 0 0 0
27 32 1 0 0 0 0
20 24 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
37 41 1 0 0 0 0
38 42 1 0 0 0 0
32 36 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
45 49 1 0 0 0 0
44 50 1 0 0 0 0
43 51 1 0 0 0 0
46 52 1 0 0 0 0
47 21 1 0 0 0 0
53 54 1 0 0 0 0
54 55 1 1 0 0 0
55 56 1 1 0 0 0
57 56 1 1 0 0 0
57 58 1 0 0 0 0
58 53 1 0 0 0 0
53 59 1 0 0 0 0
58 60 1 0 0 0 0
57 61 1 0 0 0 0
56 62 1 0 0 0 0
54 52 1 0 0 0 0
63 64 1 1 0 0 0
64 65 1 1 0 0 0
66 65 1 1 0 0 0
66 67 1 0 0 0 0
67 68 1 0 0 0 0
68 63 1 0 0 0 0
64 69 1 0 0 0 0
65 70 1 0 0 0 0
63 59 1 0 0 0 0
66 71 1 0 0 0 0
62 72 1 0 0 0 0
72 73 1 0 0 0 0
73 74 2 0 0 0 0
73 75 1 0 0 0 0
75 76 2 0 0 0 0
76 77 1 0 0 0 0
77 78 2 0 0 0 0
78 79 1 0 0 0 0
79 80 2 0 0 0 0
80 81 1 0 0 0 0
81 82 2 0 0 0 0
82 77 1 0 0 0 0
80 83 1 0 0 0 0
81 84 1 0 0 0 0
85 86 1 0 0 0 0
67 85 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 85 86
M SBL 1 1 94
M SMT 1 CH2OH
M SBV 1 94 -0.5716 -0.4834
S SKP 5
ID FL5FAGGS0017
FORMULA C53H64O33
EXACTMASS 1228.332984574
AVERAGEMASS 1229.05546
SMILES OC(C(O)1)C(Oc(c(O)3)c(O)cc(C(O5)=C(OC(C6OC(O7)C(OC(C(O)9)OC(C(O)C9O)CO)C(O)C(O)C7COC(C=Cc(c8)cc(O)c(O)c8)=O)OC(C(O)C6O)C)C(=O)c(c45)c(cc(c4)O)O)c3)OCC(OC(C2O)OC(C(C2O)O)C)1
M END