Mol:FL5FACGL0073
From Metabolomics.JP
Copyright: ARM project http://www.metabolome.jp/
66 72 0 0 0 0 0 0 0 0999 V2000
-2.7577 1.0937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7577 0.4513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2014 0.1301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6451 0.4513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6451 1.0937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2014 1.4149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0888 0.1301 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5325 0.4513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5325 1.0937 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0888 1.4149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0888 -0.3707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0236 1.4147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5905 1.0874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1575 1.4147 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1575 2.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5905 2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0236 2.0694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3138 1.4147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7243 2.3967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1384 0.0624 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2014 -0.5120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5905 3.0513 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8802 -1.9352 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.2981 -1.7850 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.3259 -1.1903 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.0352 -0.6771 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6737 -0.9024 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6173 -1.5221 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.8802 -2.4081 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2981 -2.5414 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2075 -1.1830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3074 -1.8040 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
0.6686 -1.8040 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.1278 -2.2482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2847 -2.8712 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.9236 -2.8712 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.4644 -2.4270 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9229 -2.2132 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1827 -3.1799 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2202 -0.7118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8171 -1.1149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9342 -0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7453 -0.7118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9723 -0.3186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4260 -0.3186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6988 -0.7911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2444 -0.7911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5172 -0.3186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2444 0.1539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6988 0.1539 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0617 -0.3186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7212 2.2352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1533 1.9073 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.3335 2.5378 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.1533 3.1722 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.7212 3.5002 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.5411 2.8696 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.7658 4.0105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7989 3.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9159 2.2967 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2250 -1.1093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1252 -1.5447 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0954 -3.8624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5120 -3.2791 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2173 3.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9592 2.4173 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 20 1 0 0 0 0
32 33 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
36 39 1 0 0 0 0
33 31 1 0 0 0 0
40 41 1 0 0 0 0
41 32 1 0 0 0 0
40 42 2 0 0 0 0
40 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 45 1 0 0 0 0
48 51 1 0 0 0 0
53 52 1 1 0 0 0
53 54 1 0 0 0 0
54 55 1 0 0 0 0
55 56 1 0 0 0 0
56 57 1 1 0 0 0
57 52 1 1 0 0 0
56 58 1 0 0 0 0
55 59 1 0 0 0 0
54 60 1 0 0 0 0
18 53 1 0 0 0 0
28 61 1 0 0 0 0
61 62 1 0 0 0 0
35 63 1 0 0 0 0
63 64 1 0 0 0 0
57 65 1 0 0 0 0
65 66 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 65 66
M SBL 3 1 71
M SMT 3 CH2OH
M SVB 3 71 -4.0617 3.3532
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 63 64
M SBL 2 1 69
M SMT 2 CH2OH
M SVB 2 69 1.0954 -3.8624
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 61 62
M SBL 1 1 67
M SMT 1 CH2OH
M SVB 1 67 -1.225 -1.1093
S SKP 8
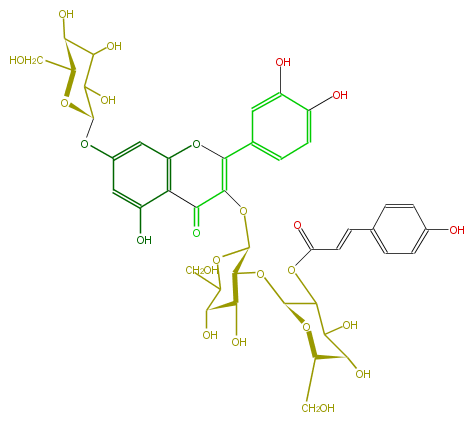
ID FL5FACGL0073
KNApSAcK_ID C00005995
NAME Quercetin 3-p-coumarylsophoroside-7-glucoside
CAS_RN 162556-36-7
FORMULA C42H46O24
EXACTMASS 934.2379024
AVERAGEMASS 934.8002399999999
SMILES O=C(c64)C(=C(c(c7)ccc(O)c(O)7)Oc(cc(cc6O)O[C@@H](O5)C(C([C@H](O)[C@@H]5CO)O)O)4)O[C@@H]([C@H]1O[C@@H](C(OC(C=Cc(c3)ccc(c3)O)=O)2)O[C@@H]([C@H](O)C2O)CO)OC([C@@H]([C@@H]1O)O)CO
M END