Mol:FL7AACGL0025
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 38 41 0 0 0 0 0 0 0 0999 V2000 | + | 38 41 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.2166 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2166 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2166 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2166 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6603 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6603 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1040 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1040 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1040 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1040 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6603 0.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6603 0.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5477 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5477 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9914 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9914 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9914 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9914 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5477 0.3250 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | -1.5477 0.3250 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4353 0.3248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4353 0.3248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1316 -0.0025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1316 -0.0025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6986 0.3248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6986 0.3248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6986 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6986 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1316 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1316 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4353 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4353 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7727 0.3248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7727 0.3248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2654 1.3068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2654 1.3068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6603 -1.6019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6603 -1.6019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5753 -1.3593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5753 -1.3593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9275 -1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9275 -1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6593 -1.8476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6593 -1.8476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1751 -1.7002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1751 -1.7002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6940 -1.8476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6940 -1.8476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9623 -1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9623 -1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4465 -1.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4465 -1.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4294 -1.5165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4294 -1.5165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5882 -1.0016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5882 -1.0016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3695 -1.3830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3695 -1.3830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1316 1.9614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1316 1.9614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2398 -0.3812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2398 -0.3812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2398 0.1493 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2398 0.1493 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6999 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6999 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3534 -0.4640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3534 -0.4640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3534 0.0463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3534 0.0463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7727 -0.8833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7727 -0.8833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1001 -1.7479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1001 -1.7479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8970 -1.9614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8970 -1.9614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 15 30 1 0 0 0 0 | + | 15 30 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 34 36 2 0 0 0 0 | + | 34 36 2 0 0 0 0 |
| − | 28 31 1 0 0 0 0 | + | 28 31 1 0 0 0 0 |
| − | 24 37 1 0 0 0 0 | + | 24 37 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 40 -6.9314 5.1979 | + | M SBV 1 40 -6.9314 5.1979 |
| − | S SKP 8 | + | S SKP 8 |
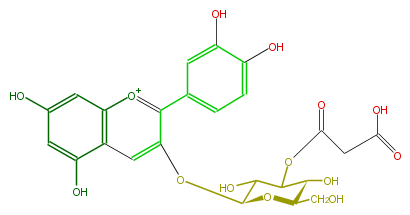
| − | ID FL7AACGL0025 | + | ID FL7AACGL0025 |
| − | KNApSAcK_ID C00006795 | + | KNApSAcK_ID C00006795 |
| − | NAME Cyanidin 3-(3''-malonylglucoside) | + | NAME Cyanidin 3-(3''-malonylglucoside) |
| − | CAS_RN 180129-51-5 | + | CAS_RN 180129-51-5 |
| − | FORMULA C24H23O14 | + | FORMULA C24H23O14 |
| − | EXACTMASS 535.108780444 | + | EXACTMASS 535.108780444 |
| − | AVERAGEMASS 535.43102 | + | AVERAGEMASS 535.43102 |
| − | SMILES C(O)C(C1O)OC(Oc(c4)c([o+1]c(c43)cc(cc3O)O)c(c2)cc(c(c2)O)O)C(O)C(OC(CC(O)=O)=O)1 | + | SMILES C(O)C(C1O)OC(Oc(c4)c([o+1]c(c43)cc(cc3O)O)c(c2)cc(c(c2)O)O)C(O)C(OC(CC(O)=O)=O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
38 41 0 0 0 0 0 0 0 0999 V2000
-3.2166 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2166 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6603 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1040 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1040 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6603 0.3250 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5477 -0.9598 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9914 -0.6386 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9914 0.0038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5477 0.3250 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
-0.4353 0.3248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1316 -0.0025 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6986 0.3248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6986 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1316 1.3069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4353 0.9795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7727 0.3248 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2654 1.3068 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6603 -1.6019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5753 -1.3593 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9275 -1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6593 -1.8476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1751 -1.7002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6940 -1.8476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9623 -1.3830 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4465 -1.5304 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4294 -1.5165 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5882 -1.0016 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3695 -1.3830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1316 1.9614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2398 -0.3812 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2398 0.1493 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6999 -0.8413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3534 -0.4640 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3534 0.0463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7727 -0.8833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1001 -1.7479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8970 -1.9614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
1 17 1 0 0 0 0
14 18 1 0 0 0 0
3 19 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
15 30 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
34 36 2 0 0 0 0
28 31 1 0 0 0 0
24 37 1 0 0 0 0
37 38 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 40
M SMT 1 CH2OH
M SBV 1 40 -6.9314 5.1979
S SKP 8
ID FL7AACGL0025
KNApSAcK_ID C00006795
NAME Cyanidin 3-(3''-malonylglucoside)
CAS_RN 180129-51-5
FORMULA C24H23O14
EXACTMASS 535.108780444
AVERAGEMASS 535.43102
SMILES C(O)C(C1O)OC(Oc(c4)c([o+1]c(c43)cc(cc3O)O)c(c2)cc(c(c2)O)O)C(O)C(OC(CC(O)=O)=O)1
M END