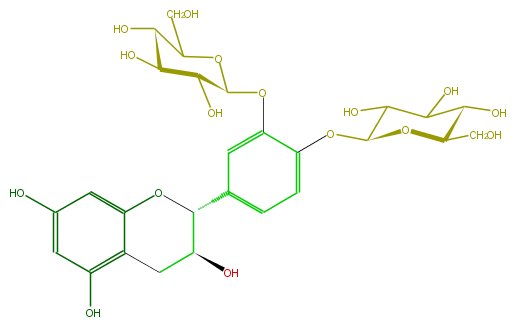
Mol:FL63ACGS0024
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.4573 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4573 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4573 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4573 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7429 -2.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7429 -2.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0284 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0284 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0284 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0284 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7429 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7429 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3140 -2.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3140 -2.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5995 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5995 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5995 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5995 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3140 -0.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3140 -0.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8851 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8851 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1647 -1.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1647 -1.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5557 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5557 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5557 0.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5557 0.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1647 0.6280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1647 0.6280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8851 0.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8851 0.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2452 0.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2452 0.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1716 -0.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1716 -0.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8851 -2.2697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8851 -2.2697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1647 1.4585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1647 1.4585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7429 -3.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7429 -3.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4093 1.1684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4093 1.1684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9944 0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9944 0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7922 0.6778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7922 0.6778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5949 0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5949 0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0099 1.1684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0099 1.1684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2120 0.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2120 0.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7432 1.0636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7432 1.0636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6657 1.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6657 1.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6513 1.0482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6513 1.0482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4012 2.7656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4012 2.7656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2958 1.9244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2958 1.9244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5139 1.8041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5139 1.8041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8941 1.4550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8941 1.4550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0832 2.1612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0832 2.1612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8120 2.1889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8120 2.1889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0215 2.7813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0215 2.7813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8789 2.2282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8789 2.2282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2126 1.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2126 1.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0842 3.0928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0842 3.0928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0987 2.5239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0987 2.5239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1863 0.5884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1863 0.5884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1716 0.0194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1716 0.0194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 6 0 0 0 | + | 9 11 1 6 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 17 14 1 0 0 0 0 | + | 17 14 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 1 0 0 0 | + | 8 19 1 1 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 17 1 0 0 0 0 | + | 23 17 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 25 42 1 0 0 0 0 | + | 25 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 0.2722 -0.9039 | + | M SBV 1 45 0.2722 -0.9039 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 42 43 | + | M SAL 2 2 42 43 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 47 -0.5914 -0.1386 | + | M SBV 2 47 -0.5914 -0.1386 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL63ACGS0024 | + | ID FL63ACGS0024 |
| − | FORMULA C27H34O16 | + | FORMULA C27H34O16 |
| − | EXACTMASS 614.18468504 | + | EXACTMASS 614.18468504 |
| − | AVERAGEMASS 614.54926 | + | AVERAGEMASS 614.54926 |
| − | SMILES c(c1C(O5)C(Cc(c54)c(O)cc(c4)O)O)c(OC(O3)C(C(C(O)C(CO)3)O)O)c(OC(O2)C(C(C(C(CO)2)O)O)O)cc1 | + | SMILES c(c1C(O5)C(Cc(c54)c(O)cc(c4)O)O)c(OC(O3)C(C(C(O)C(CO)3)O)O)c(OC(O2)C(C(C(C(CO)2)O)O)O)cc1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-4.4573 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4573 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7429 -2.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0284 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0284 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7429 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3140 -2.2697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5995 -1.8573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5995 -1.0324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3140 -0.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8851 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1647 -1.0357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5557 -0.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5557 0.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1647 0.6280 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8851 0.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2452 0.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1716 -0.6199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8851 -2.2697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1647 1.4585 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7429 -3.0928 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4093 1.1684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9944 0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7922 0.6778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5949 0.4498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0099 1.1684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2120 0.9404 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7432 1.0636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6657 1.4875 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6513 1.0482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4012 2.7656 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2958 1.9244 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5139 1.8041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8941 1.4550 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0832 2.1612 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8120 2.1889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0215 2.7813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8789 2.2282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2126 1.0393 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0842 3.0928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0987 2.5239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1863 0.5884 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1716 0.0194 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 6 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
17 14 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 1 0 0 0
15 20 1 0 0 0 0
3 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 17 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 20 1 0 0 0 0
40 41 1 0 0 0 0
36 40 1 0 0 0 0
42 43 1 0 0 0 0
25 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 0.2722 -0.9039
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 42 43
M SBL 2 1 47
M SMT 2 CH2OH
M SBV 2 47 -0.5914 -0.1386
S SKP 5
ID FL63ACGS0024
FORMULA C27H34O16
EXACTMASS 614.18468504
AVERAGEMASS 614.54926
SMILES c(c1C(O5)C(Cc(c54)c(O)cc(c4)O)O)c(OC(O3)C(C(C(O)C(CO)3)O)O)c(OC(O2)C(C(C(C(CO)2)O)O)O)cc1
M END