Mol:FL5FFCGS0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 35 38 0 0 0 0 0 0 0 0999 V2000 | + | 35 38 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0846 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0846 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0846 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0846 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5283 -0.3558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5283 -0.3558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9720 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9720 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9720 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9720 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5283 0.9289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5283 0.9289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4157 -0.3558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4157 -0.3558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1406 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1406 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1406 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1406 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4157 0.9289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4157 0.9289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4157 -0.8566 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4157 -0.8566 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6967 0.9288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6967 0.9288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2637 0.6015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2637 0.6015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8306 0.9288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8306 0.9288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8306 1.5835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8306 1.5835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2637 1.9108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2637 1.9108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6967 1.5835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6967 1.5835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6407 0.9288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6407 0.9288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5283 -0.9979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5283 -0.9979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6967 -0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6967 -0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6966 2.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6966 2.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5916 -1.5586 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.5916 -1.5586 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 2.2198 -1.9213 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2198 -1.9213 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0205 -1.2238 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.0205 -1.2238 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2198 -0.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2198 -0.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5916 -0.1593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.5916 -0.1593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 1.7909 -0.8568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.7909 -0.8568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.5612 -0.8568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5612 -0.8568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7530 -2.1608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7530 -2.1608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0682 -2.4870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0682 -2.4870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6407 -1.5819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6407 -1.5819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5471 2.4870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5471 2.4870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9884 3.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9884 3.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2503 1.5019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2503 1.5019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8138 2.4016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8138 2.4016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 15 21 1 0 0 0 0 | + | 15 21 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 20 1 0 0 0 0 | + | 26 20 1 0 0 0 0 |
| − | 16 32 1 0 0 0 0 | + | 16 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 6 34 1 0 0 0 0 | + | 6 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 37 | + | M SBL 2 1 37 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 37 -1.2503 1.5019 | + | M SVB 2 37 -1.2503 1.5019 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 32 33 | + | M SAL 1 2 32 33 |
| − | M SBL 1 1 35 | + | M SBL 1 1 35 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 35 1.5471 2.487 | + | M SVB 1 35 1.5471 2.487 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FFCGS0016 | + | ID FL5FFCGS0016 |
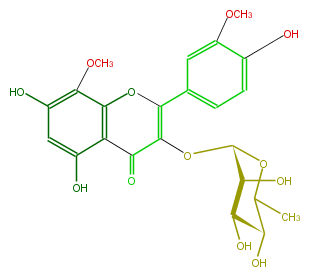
| − | KNApSAcK_ID C00005716 | + | KNApSAcK_ID C00005716 |
| − | NAME Limocitrin 3-rhamnoside | + | NAME Limocitrin 3-rhamnoside |
| − | CAS_RN 90456-56-7 | + | CAS_RN 90456-56-7 |
| − | FORMULA C23H24O12 | + | FORMULA C23H24O12 |
| − | EXACTMASS 492.126776232 | + | EXACTMASS 492.126776232 |
| − | AVERAGEMASS 492.42946 | + | AVERAGEMASS 492.42946 |
| − | SMILES O[C@H]([C@H]1OC(C(=O)4)=C(Oc(c34)c(OC)c(cc(O)3)O)c(c2)cc(OC)c(O)c2)[C@@H]([C@H](O)C(C)O1)O | + | SMILES O[C@H]([C@H]1OC(C(=O)4)=C(Oc(c34)c(OC)c(cc(O)3)O)c(c2)cc(OC)c(O)c2)[C@@H]([C@H](O)C(C)O1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
35 38 0 0 0 0 0 0 0 0999 V2000
-2.0846 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0846 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5283 -0.3558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9720 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9720 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5283 0.9289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4157 -0.3558 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1406 -0.0346 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1406 0.6077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4157 0.9289 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4157 -0.8566 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6967 0.9288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2637 0.6015 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8306 0.9288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8306 1.5835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2637 1.9108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6967 1.5835 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6407 0.9288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5283 -0.9979 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6967 -0.3557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6966 2.0835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5916 -1.5586 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
2.2198 -1.9213 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0205 -1.2238 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2198 -0.5220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5916 -0.1593 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
1.7909 -0.8568 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.5612 -0.8568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7530 -2.1608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0682 -2.4870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6407 -1.5819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5471 2.4870 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9884 3.3843 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2503 1.5019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8138 2.4016 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 0 0 0 0
15 21 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 20 1 0 0 0 0
16 32 1 0 0 0 0
32 33 1 0 0 0 0
6 34 1 0 0 0 0
34 35 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 37
M SMT 2 OCH3
M SVB 2 37 -1.2503 1.5019
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 32 33
M SBL 1 1 35
M SMT 1 OCH3
M SVB 1 35 1.5471 2.487
S SKP 8
ID FL5FFCGS0016
KNApSAcK_ID C00005716
NAME Limocitrin 3-rhamnoside
CAS_RN 90456-56-7
FORMULA C23H24O12
EXACTMASS 492.126776232
AVERAGEMASS 492.42946
SMILES O[C@H]([C@H]1OC(C(=O)4)=C(Oc(c34)c(OC)c(cc(O)3)O)c(c2)cc(OC)c(O)c2)[C@@H]([C@H](O)C(C)O1)O
M END