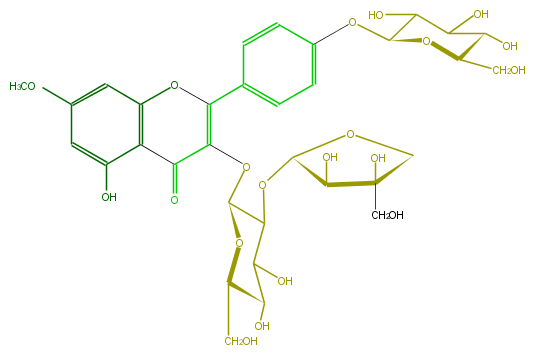
Mol:FL5FCAGL0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.8500 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8500 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8500 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8500 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1356 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1356 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4211 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4211 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4211 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4211 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1356 2.0045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1356 2.0045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7067 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7067 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9922 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9922 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9922 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9922 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7067 2.0045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7067 2.0045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7067 -0.3721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7067 -0.3721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2780 2.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2780 2.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4502 1.5840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4502 1.5840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1783 2.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1783 2.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1783 2.8452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1783 2.8452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4502 3.2655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4502 3.2655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2780 2.8452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2780 2.8452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2181 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2181 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1356 -0.3079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1356 -0.3079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9924 3.3152 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9924 3.3152 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1600 -0.8793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1600 -0.8793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5885 -0.4471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5885 -0.4471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3509 -1.2781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3509 -1.2781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5885 -2.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5885 -2.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1600 -2.5464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1600 -2.5464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0774 -1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0774 -1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1325 -0.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1325 -0.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5152 -2.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5152 -2.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0369 -3.0058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0369 -3.0058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7316 0.4971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7316 0.4971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4525 -0.0804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4525 -0.0804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4631 -0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4631 -0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2641 0.5383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2641 0.5383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9475 0.9921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9475 0.9921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4525 0.5177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4525 0.5177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4631 0.5007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4631 0.5007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3114 3.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3114 3.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7650 2.9202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7650 2.9202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5375 2.9330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5375 2.9330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2046 2.5345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2046 2.5345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7510 3.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7510 3.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9784 3.0681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9784 3.0681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5685 3.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5685 3.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6017 3.4972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6017 3.4972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2015 2.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2015 2.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5885 -3.3192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5885 -3.3192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8604 -3.4972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8604 -3.4972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5592 2.0015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5592 2.0015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2015 3.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2015 3.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9889 2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9889 2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7147 1.3009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7147 1.3009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4631 -0.7020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4631 -0.7020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4866 -0.9764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4866 -0.9764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 8 18 1 0 0 0 0 | + | 8 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 18 1 0 0 0 0 | + | 22 18 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 30 27 1 0 0 0 0 | + | 30 27 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 42 44 1 0 0 0 0 | + | 42 44 1 0 0 0 0 |
| − | 41 45 1 0 0 0 0 | + | 41 45 1 0 0 0 0 |
| − | 38 20 1 0 0 0 0 | + | 38 20 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 24 46 1 0 0 0 0 | + | 24 46 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 1 48 1 0 0 0 0 | + | 1 48 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 40 50 1 0 0 0 0 | + | 40 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 32 52 1 0 0 0 0 | + | 32 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 46 47 | + | M SAL 1 2 46 47 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 52 0.0000 1.2050 | + | M SBV 1 52 0.0000 1.2050 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 48 49 | + | M SAL 2 2 48 49 |
| − | M SBL 2 1 54 | + | M SBL 2 1 54 |
| − | M SMT 2 ^ OCH3 | + | M SMT 2 ^ OCH3 |
| − | M SBV 2 54 0.7091 -0.4094 | + | M SBV 2 54 0.7091 -0.4094 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 50 51 | + | M SAL 3 2 50 51 |
| − | M SBL 3 1 56 | + | M SBL 3 1 56 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 56 -0.7843 0.2102 | + | M SBV 3 56 -0.7843 0.2102 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 52 53 | + | M SAL 4 2 52 53 |
| − | M SBL 4 1 58 | + | M SBL 4 1 58 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SBV 4 58 0.0000 0.6629 | + | M SBV 4 58 0.0000 0.6629 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FCAGL0006 | + | ID FL5FCAGL0006 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
| − | SMILES C(O6)(C(C(C(O)C6CO)O)O)Oc(c1)ccc(C(=C(OC(C(OC(O5)C(O)C(O)(CO)C5)4)OC(C(O)C(O)4)CO)3)Oc(c2C3=O)cc(cc2O)OC)c1 | + | SMILES C(O6)(C(C(C(O)C6CO)O)O)Oc(c1)ccc(C(=C(OC(C(OC(O5)C(O)C(O)(CO)C5)4)OC(C(O)C(O)4)CO)3)Oc(c2C3=O)cc(cc2O)OC)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-3.8500 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8500 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1356 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4211 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4211 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1356 2.0045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7067 0.3545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9922 0.7670 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9922 1.5921 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7067 2.0045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7067 -0.3721 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2780 2.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4502 1.5840 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1783 2.0043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1783 2.8452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4502 3.2655 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2780 2.8452 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2181 0.3115 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1356 -0.3079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9924 3.3152 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1600 -0.8793 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5885 -0.4471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3509 -1.2781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5885 -2.1141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1600 -2.5464 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0774 -1.7154 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1325 -0.0598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5152 -2.0575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0369 -3.0058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7316 0.4971 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4525 -0.0804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4631 -0.0391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2641 0.5383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9475 0.9921 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4525 0.5177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4631 0.5007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3114 3.4665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7650 2.9202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5375 2.9330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2046 2.5345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7510 3.0808 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9784 3.0681 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5685 3.4665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6017 3.4972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2015 2.8207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5885 -3.3192 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8604 -3.4972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5592 2.0015 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2015 3.1137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9889 2.3243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7147 1.3009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4631 -0.7020 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4866 -0.9764 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
8 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 18 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 30 1 0 0 0 0
31 35 1 0 0 0 0
30 27 1 0 0 0 0
32 36 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 1 0 0 0
39 40 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 37 1 0 0 0 0
37 43 1 0 0 0 0
42 44 1 0 0 0 0
41 45 1 0 0 0 0
38 20 1 0 0 0 0
46 47 1 0 0 0 0
24 46 1 0 0 0 0
48 49 1 0 0 0 0
1 48 1 0 0 0 0
50 51 1 0 0 0 0
40 50 1 0 0 0 0
52 53 1 0 0 0 0
32 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 46 47
M SBL 1 1 52
M SMT 1 CH2OH
M SBV 1 52 0.0000 1.2050
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 48 49
M SBL 2 1 54
M SMT 2 ^ OCH3
M SBV 2 54 0.7091 -0.4094
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 50 51
M SBL 3 1 56
M SMT 3 CH2OH
M SBV 3 56 -0.7843 0.2102
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 52 53
M SBL 4 1 58
M SMT 4 CH2OH
M SBV 4 58 0.0000 0.6629
S SKP 5
ID FL5FCAGL0006
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES C(O6)(C(C(C(O)C6CO)O)O)Oc(c1)ccc(C(=C(OC(C(OC(O5)C(O)C(O)(CO)C5)4)OC(C(O)C(O)4)CO)3)Oc(c2C3=O)cc(cc2O)OC)c1
M END