Mol:FL5FAGGL0005
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1619 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1619 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1619 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1619 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6056 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6056 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0493 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0493 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0493 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0493 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6056 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6056 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5070 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5070 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0633 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0633 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0633 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0633 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5070 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5070 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5070 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5070 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7637 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7637 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3307 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3307 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8977 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8977 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8977 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8977 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3307 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3307 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7637 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7637 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6056 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6056 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5045 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5045 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8264 0.1458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8264 0.1458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5073 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5073 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3307 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3307 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4645 -0.1203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4645 -0.1203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3113 -0.0964 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.3113 -0.0964 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.9401 -0.5864 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.9401 -0.5864 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.4056 -0.3785 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.4056 -0.3785 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8898 -0.3730 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.8898 -0.3730 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.2646 0.0019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2646 0.0019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7322 -0.2449 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.7322 -0.2449 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.6372 0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6372 0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2904 -1.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2904 -1.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0993 -0.8927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0993 -0.8927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7193 -0.8883 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.7193 -0.8883 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.4185 -1.4094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.4185 -1.4094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9970 -1.2440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9970 -1.2440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5789 -1.4094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.5789 -1.4094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.8798 -0.8883 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.8798 -0.8883 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.3013 -1.0536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.3013 -1.0536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.1606 -1.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1606 -1.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3428 -0.5790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3428 -0.5790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3139 -1.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3139 -1.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7786 -1.7017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7786 -1.7017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7786 -2.5267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7786 -2.5267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9252 0.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9252 0.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8816 0.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8816 0.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 14 23 1 0 0 0 0 | + | 14 23 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 32 1 0 0 0 0 | + | 26 32 1 0 0 0 0 |
| − | 27 20 1 0 0 0 0 | + | 27 20 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 33 1 0 0 0 0 | + | 38 33 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 34 21 1 0 0 0 0 | + | 34 21 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 29 44 1 0 0 0 0 | + | 29 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 44 45 | + | M SAL 2 2 44 45 |
| − | M SBL 2 1 48 | + | M SBL 2 1 48 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 48 -3.9252 0.5899 | + | M SVB 2 48 -3.9252 0.5899 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 46 3.9319 -1.3826 | + | M SVB 1 46 3.9319 -1.3826 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGL0005 | + | ID FL5FAGGL0005 |
| − | KNApSAcK_ID C00005740 | + | KNApSAcK_ID C00005740 |
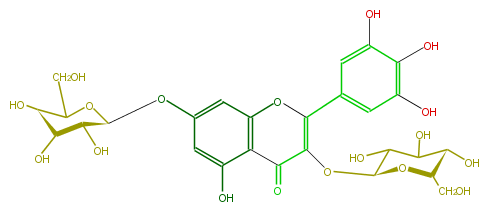
| − | NAME Myricetin 3,7-diglucoside | + | NAME Myricetin 3,7-diglucoside |
| − | CAS_RN 72078-29-6 | + | CAS_RN 72078-29-6 |
| − | FORMULA C27H30O18 | + | FORMULA C27H30O18 |
| − | EXACTMASS 642.143214156 | + | EXACTMASS 642.143214156 |
| − | AVERAGEMASS 642.5163 | + | AVERAGEMASS 642.5163 |
| − | SMILES c(c25)c(cc(c2C(C(=C(O5)c(c4)cc(c(c(O)4)O)O)O[C@@H](C(O)3)O[C@H](CO)[C@@H](C(O)3)O)=O)O)O[C@@H]([C@@H](O)1)OC(CO)[C@H](O)[C@@H]1O | + | SMILES c(c25)c(cc(c2C(C(=C(O5)c(c4)cc(c(c(O)4)O)O)O[C@@H](C(O)3)O[C@H](CO)[C@@H](C(O)3)O)=O)O)O[C@@H]([C@@H](O)1)OC(CO)[C@H](O)[C@@H]1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-1.1619 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1619 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6056 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0493 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0493 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6056 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5070 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0633 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0633 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5070 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5070 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7637 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3307 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8977 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8977 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3307 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7637 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6056 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5045 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8264 0.1458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5073 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3307 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4645 -0.1203 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3113 -0.0964 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.9401 -0.5864 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.4056 -0.3785 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8898 -0.3730 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.2646 0.0019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7322 -0.2449 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.6372 0.0223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2904 -1.0476 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0993 -0.8927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7193 -0.8883 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4185 -1.4094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9970 -1.2440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5789 -1.4094 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.8798 -0.8883 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.3013 -1.0536 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.1606 -1.0380 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3428 -0.5790 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3139 -1.1389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7786 -1.7017 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7786 -2.5267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9252 0.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8816 0.8820 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
14 23 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 29 1 0 0 0 0
29 24 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 32 1 0 0 0 0
27 20 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 33 1 0 0 0 0
33 39 1 0 0 0 0
38 40 1 0 0 0 0
37 41 1 0 0 0 0
34 21 1 0 0 0 0
36 42 1 0 0 0 0
42 43 1 0 0 0 0
29 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 44 45
M SBL 2 1 48
M SMT 2 CH2OH
M SVB 2 48 -3.9252 0.5899
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 CH2OH
M SVB 1 46 3.9319 -1.3826
S SKP 8
ID FL5FAGGL0005
KNApSAcK_ID C00005740
NAME Myricetin 3,7-diglucoside
CAS_RN 72078-29-6
FORMULA C27H30O18
EXACTMASS 642.143214156
AVERAGEMASS 642.5163
SMILES c(c25)c(cc(c2C(C(=C(O5)c(c4)cc(c(c(O)4)O)O)O[C@@H](C(O)3)O[C@H](CO)[C@@H](C(O)3)O)=O)O)O[C@@H]([C@@H](O)1)OC(CO)[C@H](O)[C@@H]1O
M END