Mol:FL5FAGGA0006
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.4771 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4771 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4771 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4771 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9208 -0.0162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9208 -0.0162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3645 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3645 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3645 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3645 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9208 1.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9208 1.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8082 -0.0162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8082 -0.0162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2519 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2519 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2519 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2519 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8082 1.2686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8082 1.2686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8082 -0.5170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8082 -0.5170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4486 1.3922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4486 1.3922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0156 1.0648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0156 1.0648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5825 1.3922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5825 1.3922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5825 2.0469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5825 2.0469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0156 2.3742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0156 2.3742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4486 2.0469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4486 2.0469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9208 -0.6583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9208 -0.6583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1894 2.3972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1894 2.3972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9720 1.2686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9720 1.2686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0156 3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0156 3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1494 1.0649 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1494 1.0649 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0928 0.1830 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.0928 0.1830 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.5352 -0.4010 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.5352 -0.4010 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2708 -0.0633 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2708 -0.0633 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9720 -0.2901 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9720 -0.2901 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.3403 0.3002 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.3403 0.3002 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.7830 0.0061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7830 0.0061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4804 -0.1706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4804 -0.1706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8455 -0.4345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8455 -0.4345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5373 -0.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5373 -0.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4678 -1.0888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4678 -1.0888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1425 -1.0888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1425 -1.0888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7388 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7388 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4555 -2.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4555 -2.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7388 -2.5393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7388 -2.5393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3053 -2.5393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3053 -2.5393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5885 -2.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5885 -2.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3053 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3053 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4562 -3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4562 -3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5878 -3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5878 -3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1536 -2.0487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1536 -2.0487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9720 -1.0147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9720 -1.0147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0442 1.0105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0442 1.0105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0113 0.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0113 0.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 14 22 1 0 0 0 0 | + | 14 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 29 8 1 0 0 0 0 | + | 29 8 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 38 42 1 0 0 0 0 | + | 38 42 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 27 44 1 0 0 0 0 | + | 27 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 44 45 | + | M SAL 1 2 44 45 |
| − | M SBL 1 1 48 | + | M SBL 1 1 48 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SVB 1 48 2.9471 -0.3705 | + | M SVB 1 48 2.9471 -0.3705 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAGGA0006 | + | ID FL5FAGGA0006 |
| − | KNApSAcK_ID C00006038 | + | KNApSAcK_ID C00006038 |
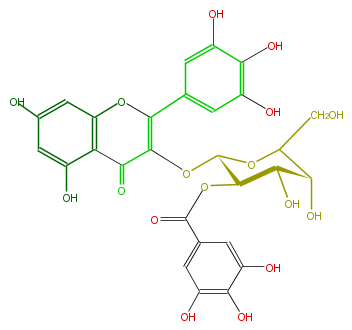
| − | NAME Myricetin 3-(2''-galloylgalactoside) | + | NAME Myricetin 3-(2''-galloylgalactoside) |
| − | CAS_RN 143222-13-3 | + | CAS_RN 143222-13-3 |
| − | FORMULA C28H24O17 | + | FORMULA C28H24O17 |
| − | EXACTMASS 632.101349342 | + | EXACTMASS 632.101349342 |
| − | AVERAGEMASS 632.47996 | + | AVERAGEMASS 632.47996 |
| − | SMILES O[C@@H]([C@H]4O)C(CO)O[C@@H]([C@@H]4OC(=O)c(c5)cc(O)c(O)c5O)OC(C2=O)=C(c(c3)cc(c(c3O)O)O)Oc(c12)cc(cc1O)O | + | SMILES O[C@@H]([C@H]4O)C(CO)O[C@@H]([C@@H]4OC(=O)c(c5)cc(O)c(O)c5O)OC(C2=O)=C(c(c3)cc(c(c3O)O)O)Oc(c12)cc(cc1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
-2.4771 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4771 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9208 -0.0162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3645 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3645 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9208 1.2686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8082 -0.0162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2519 0.3050 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2519 0.9474 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8082 1.2686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8082 -0.5170 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4486 1.3922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0156 1.0648 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5825 1.3922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5825 2.0469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0156 2.3742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4486 2.0469 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9208 -0.6583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1894 2.3972 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9720 1.2686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0156 3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1494 1.0649 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0928 0.1830 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.5352 -0.4010 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2708 -0.0633 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9720 -0.2901 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.3403 0.3002 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.7830 0.0061 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4804 -0.1706 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8455 -0.4345 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5373 -0.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4678 -1.0888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1425 -1.0888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7388 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4555 -2.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7388 -2.5393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3053 -2.5393 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5885 -2.0487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3053 -1.5581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4562 -3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5878 -3.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1536 -2.0487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9720 -1.0147 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0442 1.0105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0113 0.7560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
4 3 1 0 0 0 0
16 21 1 0 0 0 0
14 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
29 8 1 0 0 0 0
30 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 34 1 0 0 0 0
36 40 1 0 0 0 0
37 41 1 0 0 0 0
38 42 1 0 0 0 0
26 43 1 0 0 0 0
27 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 44 45
M SBL 1 1 48
M SMT 1 CH2OH
M SVB 1 48 2.9471 -0.3705
S SKP 8
ID FL5FAGGA0006
KNApSAcK_ID C00006038
NAME Myricetin 3-(2''-galloylgalactoside)
CAS_RN 143222-13-3
FORMULA C28H24O17
EXACTMASS 632.101349342
AVERAGEMASS 632.47996
SMILES O[C@@H]([C@H]4O)C(CO)O[C@@H]([C@@H]4OC(=O)c(c5)cc(O)c(O)c5O)OC(C2=O)=C(c(c3)cc(c(c3O)O)O)Oc(c12)cc(cc1O)O
M END