Mol:FL5FAFGL0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.1750 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1750 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1750 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1750 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6187 -1.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6187 -1.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0624 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0624 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0624 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0624 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6187 0.2221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6187 0.2221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4939 -1.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4939 -1.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0502 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0502 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0502 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0502 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4939 0.2221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4939 0.2221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4939 -1.5635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4939 -1.5635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6063 0.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6063 0.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1733 -0.1054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1733 -0.1054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7402 0.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7402 0.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7402 0.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7402 0.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1733 1.2040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1733 1.2040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6063 0.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6063 0.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7311 0.2220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7311 0.2220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4444 -1.1303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4444 -1.1303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6187 -1.7047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6187 -1.7047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4447 -0.3176 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -4.4447 -0.3176 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -4.0177 -0.8811 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -4.0177 -0.8811 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.4030 -0.6421 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.4030 -0.6421 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8097 -0.6357 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.8097 -0.6357 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.2408 -0.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2408 -0.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8688 -0.4300 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.8688 -0.4300 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.9219 -0.0421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9219 -0.0421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4207 -1.4117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4207 -1.4117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0507 -1.2334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0507 -1.2334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9950 -0.8733 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.9950 -0.8733 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.6941 -1.3943 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.6941 -1.3943 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.2726 -1.2290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2726 -1.2290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8546 -1.3943 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.8546 -1.3943 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 4.1555 -0.8733 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 4.1555 -0.8733 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5770 -1.0386 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.5770 -1.0386 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.4363 -1.0230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4363 -1.0230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6184 -0.5640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6184 -0.5640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5895 -1.1239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5895 -1.1239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1564 0.3303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1564 0.3303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5233 1.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5233 1.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0542 -1.6867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0542 -1.6867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0542 -2.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0542 -2.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6062 1.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6062 1.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4722 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4722 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4567 1.7801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4567 1.7801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8981 2.6774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8981 2.6774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 18 1 0 0 0 0 | + | 24 18 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 31 19 1 0 0 0 0 | + | 31 19 1 0 0 0 0 |
| − | 26 39 1 0 0 0 0 | + | 26 39 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 15 43 1 0 0 0 0 | + | 15 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 16 45 1 0 0 0 0 | + | 16 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 41 42 | + | M SAL 4 2 41 42 |
| − | M SBL 4 1 45 | + | M SBL 4 1 45 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 45 4.2076 -1.3676 | + | M SVB 4 45 4.2076 -1.3676 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 39 40 | + | M SAL 3 2 39 40 |
| − | M SBL 3 1 43 | + | M SBL 3 1 43 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 43 -4.1564 0.3303 | + | M SVB 3 43 -4.1564 0.3303 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 45 46 | + | M SAL 2 2 45 46 |
| − | M SBL 2 1 49 | + | M SBL 2 1 49 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 49 2.4567 1.7801 | + | M SVB 2 49 2.4567 1.7801 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 43 44 | + | M SAL 1 2 43 44 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 47 3.307 1.2039 | + | M SVB 1 47 3.307 1.2039 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAFGL0003 | + | ID FL5FAFGL0003 |
| − | KNApSAcK_ID C00005621 | + | KNApSAcK_ID C00005621 |
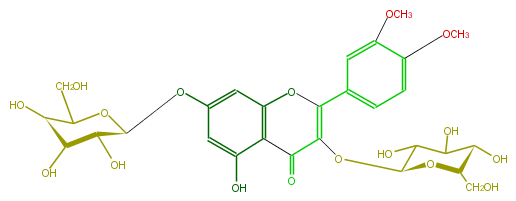
| − | NAME Dillenetin 3,7-diglucoside;Quercetin 3',4'-dimethyl ether 3,7-diglucoside;2-(3,4-Dimethoxyphenyl)-3,7-bis(beta-D-glucopyranosyloxy)-5-hydroxy-4H-1-benzopyran-4-one | + | NAME Dillenetin 3,7-diglucoside;Quercetin 3',4'-dimethyl ether 3,7-diglucoside;2-(3,4-Dimethoxyphenyl)-3,7-bis(beta-D-glucopyranosyloxy)-5-hydroxy-4H-1-benzopyran-4-one |
| − | CAS_RN 141852-52-0 | + | CAS_RN 141852-52-0 |
| − | FORMULA C29H34O17 | + | FORMULA C29H34O17 |
| − | EXACTMASS 654.179599662 | + | EXACTMASS 654.179599662 |
| − | AVERAGEMASS 654.57006 | + | AVERAGEMASS 654.57006 |
| − | SMILES c(c43)(cc(O[C@H](O5)[C@@H](O)[C@@H](O)[C@@H](O)C(CO)5)cc4O)OC(=C(C3=O)O[C@H](O2)C(O)C([C@@H](O)[C@H]2CO)O)c(c1)ccc(c(OC)1)OC | + | SMILES c(c43)(cc(O[C@H](O5)[C@@H](O)[C@@H](O)[C@@H](O)C(CO)5)cc4O)OC(=C(C3=O)O[C@H](O2)C(O)C([C@@H](O)[C@H]2CO)O)c(c1)ccc(c(OC)1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.1750 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1750 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6187 -1.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0624 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0624 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6187 0.2221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4939 -1.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0502 -0.7414 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0502 -0.0991 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4939 0.2221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4939 -1.5635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6063 0.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1733 -0.1054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7402 0.2220 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7402 0.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1733 1.2040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6063 0.8767 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7311 0.2220 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4444 -1.1303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6187 -1.7047 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4447 -0.3176 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-4.0177 -0.8811 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.4030 -0.6421 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8097 -0.6357 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.2408 -0.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8688 -0.4300 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.9219 -0.0421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4207 -1.4117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0507 -1.2334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9950 -0.8733 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.6941 -1.3943 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.2726 -1.2290 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8546 -1.3943 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
4.1555 -0.8733 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5770 -1.0386 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.4363 -1.0230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6184 -0.5640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5895 -1.1239 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1564 0.3303 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5233 1.1044 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0542 -1.6867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0542 -2.5116 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6062 1.3768 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4722 1.8768 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4567 1.7801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8981 2.6774 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 18 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
31 19 1 0 0 0 0
26 39 1 0 0 0 0
39 40 1 0 0 0 0
33 41 1 0 0 0 0
41 42 1 0 0 0 0
15 43 1 0 0 0 0
43 44 1 0 0 0 0
16 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 41 42
M SBL 4 1 45
M SMT 4 CH2OH
M SVB 4 45 4.2076 -1.3676
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 39 40
M SBL 3 1 43
M SMT 3 CH2OH
M SVB 3 43 -4.1564 0.3303
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 45 46
M SBL 2 1 49
M SMT 2 OCH3
M SVB 2 49 2.4567 1.7801
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 43 44
M SBL 1 1 47
M SMT 1 OCH3
M SVB 1 47 3.307 1.2039
S SKP 8
ID FL5FAFGL0003
KNApSAcK_ID C00005621
NAME Dillenetin 3,7-diglucoside;Quercetin 3',4'-dimethyl ether 3,7-diglucoside;2-(3,4-Dimethoxyphenyl)-3,7-bis(beta-D-glucopyranosyloxy)-5-hydroxy-4H-1-benzopyran-4-one
CAS_RN 141852-52-0
FORMULA C29H34O17
EXACTMASS 654.179599662
AVERAGEMASS 654.57006
SMILES c(c43)(cc(O[C@H](O5)[C@@H](O)[C@@H](O)[C@@H](O)C(CO)5)cc4O)OC(=C(C3=O)O[C@H](O2)C(O)C([C@@H](O)[C@H]2CO)O)c(c1)ccc(c(OC)1)OC
M END