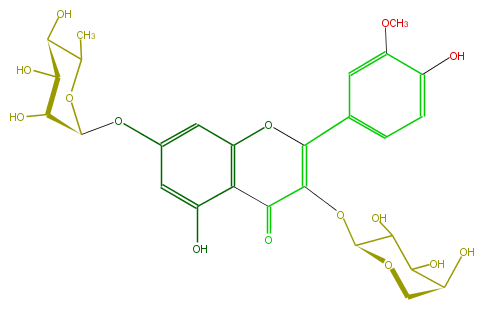
Mol:FL5FADGS0013
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6211 -0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6211 -0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6211 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6211 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9063 -1.5270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9063 -1.5270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1914 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1914 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1915 -0.2889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1915 -0.2889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9063 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9063 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5233 -1.5270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5233 -1.5270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2380 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2380 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2381 -0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2381 -0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5233 0.1237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5233 0.1237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5235 -2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5235 -2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1383 0.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1383 0.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8668 -0.1380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8668 -0.1380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5953 0.2826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5953 0.2826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5953 1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5953 1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8668 1.5445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8668 1.5445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1382 1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1382 1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9062 -2.3520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9062 -2.3520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2351 1.5174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2351 1.5174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4749 0.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4749 0.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9750 -1.6655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9750 -1.6655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0004 -2.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0004 -2.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2672 -2.2979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2672 -2.2979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9308 -2.6666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9308 -2.6666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3025 -3.3332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3025 -3.3332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0358 -3.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0358 -3.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3722 -2.7682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3722 -2.7682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6709 -1.7304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6709 -1.7304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8354 -2.6527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8354 -2.6527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4331 -2.4124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4331 -2.4124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9055 0.3134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9055 0.3134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2307 -0.0762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2307 -0.0762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4449 0.6731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4449 0.6731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2307 1.4269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2307 1.4269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9055 1.8166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9055 1.8166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6914 1.0673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6914 1.0673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6296 2.3617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6296 2.3617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2400 1.9354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2400 1.9354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2985 1.2299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2985 1.2299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4331 0.2830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4331 0.2830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8611 2.1802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8611 2.1802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4283 3.3332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4283 3.3332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 31 40 1 0 0 0 0 | + | 31 40 1 0 0 0 0 |
| − | 32 20 1 0 0 0 0 | + | 32 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 16 41 1 0 0 0 0 | + | 16 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 46 0.0057 -0.6357 | + | M SBV 1 46 0.0057 -0.6357 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGS0013 | + | ID FL5FADGS0013 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES O(c(c2)cc(O)c(C3=O)c(OC(c(c5)ccc(c5OC)O)=C3OC(C4O)OCC(C4O)O)2)C(C(O)1)OC(C)C(O)C1O | + | SMILES O(c(c2)cc(O)c(C3=O)c(OC(c(c5)ccc(c5OC)O)=C3OC(C4O)OCC(C4O)O)2)C(C(O)1)OC(C)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-1.6211 -0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6211 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9063 -1.5270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1914 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1915 -0.2889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9063 0.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5233 -1.5270 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2380 -1.1143 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2381 -0.2890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5233 0.1237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5235 -2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1383 0.2827 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8668 -0.1380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5953 0.2826 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5953 1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8668 1.5445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1382 1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9062 -2.3520 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2351 1.5174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4749 0.2040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9750 -1.6655 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0004 -2.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2672 -2.2979 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9308 -2.6666 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3025 -3.3332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0358 -3.1369 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3722 -2.7682 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6709 -1.7304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8354 -2.6527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4331 -2.4124 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9055 0.3134 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2307 -0.0762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4449 0.6731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2307 1.4269 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9055 1.8166 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6914 1.0673 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6296 2.3617 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2400 1.9354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2985 1.2299 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4331 0.2830 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8611 2.1802 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4283 3.3332 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
21 23 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
31 40 1 0 0 0 0
32 20 1 0 0 0 0
41 42 1 0 0 0 0
16 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 OCH3
M SBV 1 46 0.0057 -0.6357
S SKP 5
ID FL5FADGS0013
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES O(c(c2)cc(O)c(C3=O)c(OC(c(c5)ccc(c5OC)O)=C3OC(C4O)OCC(C4O)O)2)C(C(O)1)OC(C)C(O)C1O
M END