Mol:FL5FADGL0023
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.9924 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9924 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9924 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9924 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2778 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2778 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5633 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5633 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5633 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5633 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2778 0.4992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2778 0.4992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1514 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1514 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8659 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8659 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8659 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8659 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1514 0.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1514 0.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1514 -1.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1514 -1.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5593 0.5136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5593 0.5136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2876 0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2876 0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0159 0.5136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0159 0.5136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0159 1.3546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0159 1.3546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2876 1.7751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2876 1.7751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5593 1.3546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5593 1.3546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2778 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2778 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7482 1.7773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7482 1.7773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8459 0.5795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8459 0.5795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6850 -1.2687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6850 -1.2687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6275 -0.8372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6275 -0.8372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2412 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2412 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9842 -1.2941 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9842 -1.2941 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7317 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7317 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1181 -0.8372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1181 -0.8372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3751 -1.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3751 -1.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0144 -0.8452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0144 -0.8452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9660 -0.4207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9660 -0.4207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9048 -1.0051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9048 -1.0051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4828 -1.3221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4828 -1.3221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9827 -1.9871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9827 -1.9871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4082 -2.5161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4082 -2.5161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0534 -3.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0534 -3.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8578 -3.0466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8578 -3.0466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3862 -2.5345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3862 -2.5345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4041 -2.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4041 -2.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0534 -3.6668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0534 -3.6668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8578 -3.6685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8578 -3.6685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2698 0.7507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2698 0.7507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5952 0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5952 0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8093 1.1102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8093 1.1102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5952 1.8637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5952 1.8637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2698 2.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2698 2.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0558 1.5043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0558 1.5043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9747 2.8614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9747 2.8614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6059 2.4973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6059 2.4973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7438 1.9749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7438 1.9749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9827 0.7410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9827 0.7410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2876 2.5159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2876 2.5159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8545 3.6685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8545 3.6685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0534 -2.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0534 -2.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2725 -1.5713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2725 -1.5713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 36 35 1 1 0 0 0 | + | 36 35 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 33 1 0 0 0 0 | + | 37 33 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 35 39 1 0 0 0 0 | + | 35 39 1 0 0 0 0 |
| − | 36 32 1 0 0 0 0 | + | 36 32 1 0 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 40 1 1 0 0 0 | + | 45 40 1 1 0 0 0 |
| − | 44 46 1 0 0 0 0 | + | 44 46 1 0 0 0 0 |
| − | 43 47 1 0 0 0 0 | + | 43 47 1 0 0 0 0 |
| − | 45 48 1 0 0 0 0 | + | 45 48 1 0 0 0 0 |
| − | 40 49 1 0 0 0 0 | + | 40 49 1 0 0 0 0 |
| − | 41 20 1 0 0 0 0 | + | 41 20 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 16 50 1 0 0 0 0 | + | 16 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 34 52 1 0 0 0 0 | + | 34 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 56 0.0000 -0.7408 | + | M SBV 1 56 0.0000 -0.7408 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 58 0.0000 -0.4967 | + | M SBV 2 58 0.0000 -0.4967 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0023 | + | ID FL5FADGL0023 |
| − | FORMULA C33H40O20 | + | FORMULA C33H40O20 |
| − | EXACTMASS 756.21129372 | + | EXACTMASS 756.21129372 |
| − | AVERAGEMASS 756.6587 | + | AVERAGEMASS 756.6587 |
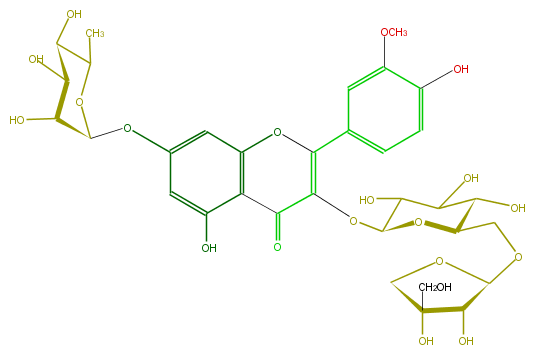
| − | SMILES OCC(C6)(C(O)C(O6)OCC(C(O)5)OC(C(O)C5O)OC(C(=O)1)=C(c(c4)cc(c(O)c4)OC)Oc(c2)c1c(O)cc2OC(O3)C(O)C(O)C(C3C)O)O | + | SMILES OCC(C6)(C(O)C(O6)OCC(C(O)5)OC(C(O)C5O)OC(C(=O)1)=C(c(c4)cc(c(O)c4)OC)Oc(c2)c1c(O)cc2OC(O3)C(O)C(O)C(C3C)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-1.9924 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9924 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2778 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5633 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5633 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2778 0.4992 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1514 -1.1509 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8659 -0.7383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8659 0.0867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1514 0.4992 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1514 -1.7943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5593 0.5136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2876 0.0932 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0159 0.5136 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0159 1.3546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2876 1.7751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5593 1.3546 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2778 -1.8182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7482 1.7773 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8459 0.5795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6850 -1.2687 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6275 -0.8372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2412 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9842 -1.2941 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7317 -1.5064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1181 -0.8372 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3751 -1.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0144 -0.8452 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9660 -0.4207 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9048 -1.0051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4828 -1.3221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9827 -1.9871 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4082 -2.5161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0534 -3.0879 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8578 -3.0466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3862 -2.5345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4041 -2.0772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0534 -3.6668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8578 -3.6685 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2698 0.7507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5952 0.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8093 1.1102 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5952 1.8637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2698 2.2533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0558 1.5043 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9747 2.8614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6059 2.4973 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7438 1.9749 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9827 0.7410 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2876 2.5159 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8545 3.6685 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0534 -2.5911 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2725 -1.5713 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
25 31 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 0 0 0 0
33 34 1 1 0 0 0
34 35 1 1 0 0 0
36 35 1 1 0 0 0
36 37 1 0 0 0 0
37 33 1 0 0 0 0
34 38 1 0 0 0 0
35 39 1 0 0 0 0
36 32 1 0 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 40 1 1 0 0 0
44 46 1 0 0 0 0
43 47 1 0 0 0 0
45 48 1 0 0 0 0
40 49 1 0 0 0 0
41 20 1 0 0 0 0
50 51 1 0 0 0 0
16 50 1 0 0 0 0
52 53 1 0 0 0 0
34 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 OCH3
M SBV 1 56 0.0000 -0.7408
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 CH2OH
M SBV 2 58 0.0000 -0.4967
S SKP 5
ID FL5FADGL0023
FORMULA C33H40O20
EXACTMASS 756.21129372
AVERAGEMASS 756.6587
SMILES OCC(C6)(C(O)C(O6)OCC(C(O)5)OC(C(O)C5O)OC(C(=O)1)=C(c(c4)cc(c(O)c4)OC)Oc(c2)c1c(O)cc2OC(O3)C(O)C(O)C(C3C)O)O
M END