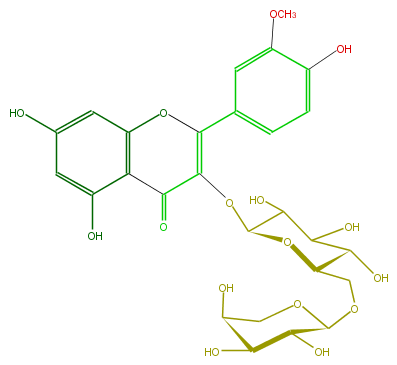
Mol:FL5FADGL0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8525 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8525 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8525 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8525 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1377 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1377 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4231 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4231 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4231 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4231 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1377 0.8262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1377 0.8262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7083 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7083 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0063 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0063 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0063 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0063 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7083 0.8262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7083 0.8262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7083 -1.4679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7083 -1.4679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7207 0.8261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7207 0.8261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4493 0.4054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4493 0.4054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1777 0.8261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1777 0.8261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1777 1.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1777 1.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4493 2.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4493 2.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7207 1.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7207 1.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5669 0.8261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5669 0.8261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1377 -1.6494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1377 -1.6494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8380 2.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8380 2.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6839 -1.1708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6839 -1.1708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9868 -1.5732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9868 -1.5732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7675 -1.7686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7675 -1.7686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3313 -2.3494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3313 -2.3494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0283 -1.9471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0283 -1.9471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2476 -1.7516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2476 -1.7516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2267 -0.9272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2267 -0.9272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9929 -1.4692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9929 -1.4692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5669 -2.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5669 -2.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9979 -2.5420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9979 -2.5420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4548 -3.2849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4548 -3.2849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0643 -3.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0643 -3.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8242 -3.5837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8242 -3.5837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5825 -3.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5825 -3.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9829 -3.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9829 -3.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2044 -3.3644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2044 -3.3644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4772 -2.6778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4772 -2.6778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3413 -3.9521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3413 -3.9521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3835 -3.9519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3835 -3.9519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1201 -3.1027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1201 -3.1027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6032 -0.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6032 -0.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4847 2.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4847 2.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0518 3.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0518 3.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | 41 8 1 0 0 0 0 | + | 41 8 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 47 | + | M SBL 1 1 47 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 47 -0.0354 -0.7182 | + | M SBV 1 47 -0.0354 -0.7182 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0002 | + | ID FL5FADGL0002 |
| − | FORMULA C27H30O16 | + | FORMULA C27H30O16 |
| − | EXACTMASS 610.153384912 | + | EXACTMASS 610.153384912 |
| − | AVERAGEMASS 610.5175 | + | AVERAGEMASS 610.5175 |
| − | SMILES C(O)(C(O)1)C(O)C(OC(C4=O)=C(Oc(c5)c(c(O)cc5O)4)c(c3)ccc(c3OC)O)OC(COC(O2)C(O)C(C(O)C2)O)1 | + | SMILES C(O)(C(O)1)C(O)C(OC(C4=O)=C(Oc(c5)c(c(O)cc5O)4)c(c3)ccc(c3OC)O)OC(COC(O2)C(O)C(C(O)C2)O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
-2.8525 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8525 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1377 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4231 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4231 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1377 0.8262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7083 -0.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0063 -0.4116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0063 0.4135 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7083 0.8262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7083 -1.4679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7207 0.8261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4493 0.4054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1777 0.8261 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1777 1.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4493 2.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7207 1.6672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5669 0.8261 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1377 -1.6494 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8380 2.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6839 -1.1708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9868 -1.5732 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7675 -1.7686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3313 -2.3494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0283 -1.9471 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2476 -1.7516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2267 -0.9272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9929 -1.4692 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5669 -2.4858 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9979 -2.5420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4548 -3.2849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0643 -3.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8242 -3.5837 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5825 -3.5092 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9829 -3.0055 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2044 -3.3644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4772 -2.6778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3413 -3.9521 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3835 -3.9519 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1201 -3.1027 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6032 -0.9727 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4847 2.8059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0518 3.9587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
40 30 1 0 0 0 0
22 41 1 0 0 0 0
41 8 1 0 0 0 0
42 43 1 0 0 0 0
16 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 47
M SMT 1 OCH3
M SBV 1 47 -0.0354 -0.7182
S SKP 5
ID FL5FADGL0002
FORMULA C27H30O16
EXACTMASS 610.153384912
AVERAGEMASS 610.5175
SMILES C(O)(C(O)1)C(O)C(OC(C4=O)=C(Oc(c5)c(c(O)cc5O)4)c(c3)ccc(c3OC)O)OC(COC(O2)C(O)C(C(O)C2)O)1
M END