Mol:FL5FADGA0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 45 49 0 0 0 0 0 0 0 0999 V2000 | + | 45 49 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.1607 -4.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1607 -4.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7811 -4.3782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7811 -4.3782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2353 -3.9240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2353 -3.9240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0691 -3.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0691 -3.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4486 -3.1373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4486 -3.1373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0056 -3.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0056 -3.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5233 -2.8493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5233 -2.8493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3570 -2.2289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3570 -2.2289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7366 -2.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7366 -2.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2824 -2.5168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2824 -2.5168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0070 -2.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0070 -2.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4882 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4882 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9512 -0.8079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9512 -0.8079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7817 -0.1755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7817 -0.1755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1494 -0.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1494 -0.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3136 -0.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3136 -0.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1441 -1.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1441 -1.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8555 -4.0901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8555 -4.0901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1094 0.9599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1094 0.9599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3819 -4.7545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3819 -4.7545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9008 -1.9149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9008 -1.9149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4355 -0.8653 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.4355 -0.8653 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.1401 -1.3769 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.1401 -1.3769 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7082 -1.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7082 -1.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2796 -1.3769 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.2796 -1.3769 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.5751 -0.8653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.5751 -0.8653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0070 -1.0276 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.0070 -1.0276 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2414 -0.3317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2414 -0.3317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0478 -0.5616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0478 -0.5616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5751 -0.1572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5751 -0.1572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7906 -6.8972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -0.7906 -6.8972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -0.2214 -6.6655 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2214 -6.6655 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.2837 -6.0954 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.2837 -6.0954 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.1556 -5.5958 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.1556 -5.5958 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.6148 -5.8608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6148 -5.8608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4974 -6.3763 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -0.4974 -6.3763 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -0.9936 -7.4547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9936 -7.4547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1336 -7.1232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1336 -7.1232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2921 -5.9326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2921 -5.9326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7500 -1.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7500 -1.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4645 -1.7739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4645 -1.7739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7967 -0.0461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7967 -0.0461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5492 0.6125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5492 0.6125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2005 -6.9047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2005 -6.9047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0222 -6.9766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0222 -6.9766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 21 23 1 0 0 0 0 | + | 21 23 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 44 45 | + | M SAL 3 2 44 45 |
| − | M SBL 3 1 48 | + | M SBL 3 1 48 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 48 -3.77 0.661 | + | M SVB 3 48 -3.77 0.661 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 40 41 | + | M SAL 2 2 40 41 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 44 3.75 -1.3614 | + | M SVB 2 44 3.75 -1.3614 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 42 43 | + | M SAL 1 2 42 43 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 46 2.6654 1.8043 | + | M SVB 1 46 2.6654 1.8043 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGA0008 | + | ID FL5FADGA0008 |
| − | KNApSAcK_ID C00005552 | + | KNApSAcK_ID C00005552 |
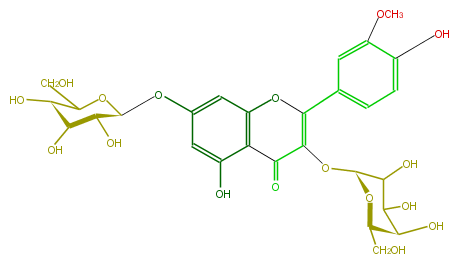
| − | NAME Isorhamnetin 3-galactoside-7-glucoside | + | NAME Isorhamnetin 3-galactoside-7-glucoside |
| − | CAS_RN 33906-93-3 | + | CAS_RN 33906-93-3 |
| − | FORMULA C28H32O17 | + | FORMULA C28H32O17 |
| − | EXACTMASS 640.163949598 | + | EXACTMASS 640.163949598 |
| − | AVERAGEMASS 640.54348 | + | AVERAGEMASS 640.54348 |
| − | SMILES c(c1)(O)c(C3=O)c(OC(c(c5)cc(OC)c(O)c5)=C3O[C@H](O4)C(C([C@@H](O)[C@H]4CO)O)O)cc(O[C@H](O2)[C@@H](O)[C@@H](O)[C@@H](O)C(CO)2)1 | + | SMILES c(c1)(O)c(C3=O)c(OC(c(c5)cc(OC)c(O)c5)=C3O[C@H](O4)C(C([C@@H](O)[C@H]4CO)O)O)cc(O[C@H](O2)[C@@H](O)[C@@H](O)[C@@H](O)C(CO)2)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
45 49 0 0 0 0 0 0 0 0999 V2000
0.1607 -4.2120 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7811 -4.3782 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2353 -3.9240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0691 -3.3035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4486 -3.1373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0056 -3.5915 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5233 -2.8493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3570 -2.2289 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7366 -2.0626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2824 -2.5168 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0070 -2.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4882 -1.2708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9512 -0.8079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7817 -0.1755 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1494 -0.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3136 -0.4690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1441 -1.1014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8555 -4.0901 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1094 0.9599 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3819 -4.7545 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9008 -1.9149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4355 -0.8653 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.1401 -1.3769 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7082 -1.2146 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2796 -1.3769 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.5751 -0.8653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0070 -1.0276 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2414 -0.3317 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0478 -0.5616 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5751 -0.1572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7906 -6.8972 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-0.2214 -6.6655 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.2837 -6.0954 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.1556 -5.5958 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.6148 -5.8608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4974 -6.3763 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-0.9936 -7.4547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1336 -7.1232 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2921 -5.9326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7500 -1.3614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4645 -1.7739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7967 -0.0461 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5492 0.6125 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2005 -6.9047 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0222 -6.9766 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
21 23 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 20 1 0 0 0 0
25 40 1 0 0 0 0
40 41 1 0 0 0 0
16 42 1 0 0 0 0
42 43 1 0 0 0 0
36 44 1 0 0 0 0
44 45 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 44 45
M SBL 3 1 48
M SMT 3 CH2OH
M SVB 3 48 -3.77 0.661
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 40 41
M SBL 2 1 44
M SMT 2 CH2OH
M SVB 2 44 3.75 -1.3614
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 42 43
M SBL 1 1 46
M SMT 1 OCH3
M SVB 1 46 2.6654 1.8043
S SKP 8
ID FL5FADGA0008
KNApSAcK_ID C00005552
NAME Isorhamnetin 3-galactoside-7-glucoside
CAS_RN 33906-93-3
FORMULA C28H32O17
EXACTMASS 640.163949598
AVERAGEMASS 640.54348
SMILES c(c1)(O)c(C3=O)c(OC(c(c5)cc(OC)c(O)c5)=C3O[C@H](O4)C(C([C@@H](O)[C@H]4CO)O)O)cc(O[C@H](O2)[C@@H](O)[C@@H](O)[C@@H](O)C(CO)2)1
M END