Mol:FL5FACGA0032
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.9512 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9512 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9512 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9512 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3949 0.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3949 0.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8386 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8386 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8386 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8386 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3949 1.7815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3949 1.7815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2822 0.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2822 0.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7259 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7259 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7259 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7259 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2822 1.7815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2822 1.7815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2822 -0.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2822 -0.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1699 1.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1699 1.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6029 1.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6029 1.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0359 1.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0359 1.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0359 2.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0359 2.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6029 2.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6029 2.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1699 2.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1699 2.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0778 0.2677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0778 0.2677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0324 0.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0324 0.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4201 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4201 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3254 0.3007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3254 0.3007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0754 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0754 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4631 0.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4631 0.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7176 0.5461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7176 0.5461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7595 0.5643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7595 0.5643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9864 1.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9864 1.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4631 1.5223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4631 1.5223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5309 2.7633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5309 2.7633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3949 -0.1453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3949 -0.1453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5621 1.6588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5621 1.6588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7043 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7043 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0499 -0.5109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0499 -0.5109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0507 -1.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0507 -1.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3949 -1.6202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3949 -1.6202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7932 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7932 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6029 3.4179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6029 3.4179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7932 -2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7932 -2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3829 -2.7373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3829 -2.7373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9726 -2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9726 -2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9726 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9726 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3829 -1.3754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3829 -1.3754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5621 -2.7372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5621 -2.7372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3829 -3.4179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3829 -3.4179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5621 -1.3756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5621 -1.3756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 1 30 1 0 0 0 0 | + | 1 30 1 0 0 0 0 |
| − | 22 31 1 0 0 0 0 | + | 22 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 16 36 1 0 0 0 0 | + | 16 36 1 0 0 0 0 |
| − | 35 37 2 0 0 0 0 | + | 35 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 35 1 0 0 0 0 | + | 41 35 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 38 43 1 0 0 0 0 | + | 38 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGA0032 | + | ID FL5FACGA0032 |
| − | KNApSAcK_ID C00005951 | + | KNApSAcK_ID C00005951 |
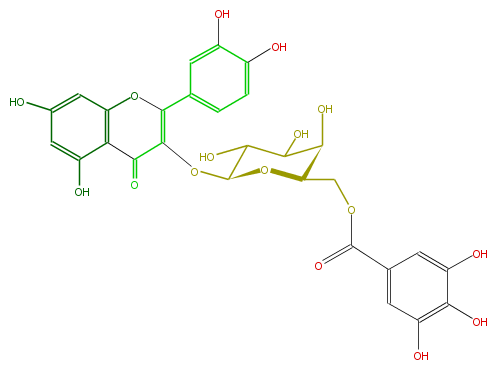
| − | NAME Quercetin 3-(6''-galloylgalactoside) | + | NAME Quercetin 3-(6''-galloylgalactoside) |
| − | CAS_RN 53171-28-1 | + | CAS_RN 53171-28-1 |
| − | FORMULA C28H24O16 | + | FORMULA C28H24O16 |
| − | EXACTMASS 616.1064347199999 | + | EXACTMASS 616.1064347199999 |
| − | AVERAGEMASS 616.48056 | + | AVERAGEMASS 616.48056 |
| − | SMILES c(c1)(O)cc(c(C(=O)3)c1OC(=C(OC(C5O)OC(C(O)C5O)COC(=O)c(c4)cc(O)c(c(O)4)O)3)c(c2)ccc(c2O)O)O | + | SMILES c(c1)(O)cc(c(C(=O)3)c1OC(=C(OC(C5O)OC(C(O)C5O)COC(=O)c(c4)cc(O)c(c(O)4)O)3)c(c2)ccc(c2O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
-3.9512 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9512 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3949 0.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8386 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8386 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3949 1.7815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2822 0.4968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7259 0.8180 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7259 1.4603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2822 1.7815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2822 -0.0040 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1699 1.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6029 1.4541 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0359 1.7814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0359 2.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6029 2.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1699 2.4361 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0778 0.2677 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0324 0.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4201 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3254 0.3007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0754 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4631 0.7591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7176 0.5461 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7595 0.5643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9864 1.0117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4631 1.5223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5309 2.7633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3949 -0.1453 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5621 1.6588 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7043 0.0877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0499 -0.5109 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0507 -1.2415 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3949 -1.6202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7932 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6029 3.4179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7932 -2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3829 -2.7373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9726 -2.3968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9726 -1.7159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3829 -1.3754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5621 -2.7372 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3829 -3.4179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5621 -1.3756 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
1 30 1 0 0 0 0
22 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
16 36 1 0 0 0 0
35 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 35 1 0 0 0 0
39 42 1 0 0 0 0
38 43 1 0 0 0 0
40 44 1 0 0 0 0
S SKP 8
ID FL5FACGA0032
KNApSAcK_ID C00005951
NAME Quercetin 3-(6''-galloylgalactoside)
CAS_RN 53171-28-1
FORMULA C28H24O16
EXACTMASS 616.1064347199999
AVERAGEMASS 616.48056
SMILES c(c1)(O)cc(c(C(=O)3)c1OC(=C(OC(C5O)OC(C(O)C5O)COC(=O)c(c4)cc(O)c(c(O)4)O)3)c(c2)ccc(c2O)O)O
M END