Mol:FL5FABGI0012
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 59 64 0 0 0 0 0 0 0 0999 V2000 | + | 59 64 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4428 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4428 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4428 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4428 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7280 0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7280 0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0132 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0132 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0132 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0132 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7280 2.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7280 2.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7015 0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7015 0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4163 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4163 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4163 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4163 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7015 2.1185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7015 2.1185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7053 -0.3082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7053 -0.3082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1308 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1308 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8593 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8593 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5878 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5878 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5878 2.9594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5878 2.9594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8593 3.3801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8593 3.3801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1308 2.9594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1308 2.9594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1573 2.1182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1573 2.1182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9228 0.3807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9228 0.3807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7280 -0.3573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7280 -0.3573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7280 2.9435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7280 2.9435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4425 3.3561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4425 3.3561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4425 4.1811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4425 4.1811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7280 4.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7280 4.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1570 4.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1570 4.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5999 2.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5999 2.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1230 1.5966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1230 1.5966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4362 1.8637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4362 1.8637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7736 1.8709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7736 1.8709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2552 2.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2552 2.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8560 2.0354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8560 2.0354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2341 2.0231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2341 2.0231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8387 1.6613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8387 1.6613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6289 1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6289 1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2659 -0.8352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2659 -0.8352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2659 -1.4928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2659 -1.4928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7230 -1.0202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7230 -1.0202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3642 -0.8586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3642 -0.8586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3643 -0.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3643 -0.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9070 -0.6737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9070 -0.6737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5444 -1.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5444 -1.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7614 -1.5181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7614 -1.5181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7309 -1.1896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7309 -1.1896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5573 -1.3962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5573 -1.3962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4741 -2.1694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4741 -2.1694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8047 -2.5559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8047 -2.5559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5546 -2.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5546 -2.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0959 -3.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0959 -3.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7654 -2.9150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7654 -2.9150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0155 -2.7273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0155 -2.7273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0769 -1.8091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0769 -1.8091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7945 -2.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7945 -2.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0396 -3.2254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0396 -3.2254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3161 3.3800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3161 3.3800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2341 2.8500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2341 2.8500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6687 -3.6754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6687 -3.6754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1387 -4.5935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1387 -4.5935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4143 2.5659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4143 2.5659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7769 3.5619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7769 3.5619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 6 21 1 0 0 0 0 | + | 6 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 2 0 0 0 0 | + | 22 23 2 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 23 25 1 0 0 0 0 | + | 23 25 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 29 28 1 1 0 0 0 | + | 29 28 1 1 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 26 1 0 0 0 0 | + | 31 26 1 0 0 0 0 |
| − | 26 32 1 0 0 0 0 | + | 26 32 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 18 1 0 0 0 0 | + | 29 18 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 35 1 0 0 0 0 | + | 40 35 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | 39 19 1 0 0 0 0 | + | 39 19 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 47 48 1 1 0 0 0 | + | 47 48 1 1 0 0 0 |
| − | 49 48 1 1 0 0 0 | + | 49 48 1 1 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 50 52 1 0 0 0 0 | + | 50 52 1 0 0 0 0 |
| − | 49 53 1 0 0 0 0 | + | 49 53 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 15 54 1 0 0 0 0 | + | 15 54 1 0 0 0 0 |
| − | 56 57 1 0 0 0 0 | + | 56 57 1 0 0 0 0 |
| − | 48 56 1 0 0 0 0 | + | 48 56 1 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 31 58 1 0 0 0 0 | + | 31 58 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 60 -0.7283 -0.4205 | + | M SBV 1 60 -0.7283 -0.4205 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 56 57 | + | M SAL 2 2 56 57 |
| − | M SBL 2 1 62 | + | M SBL 2 1 62 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 62 -0.5728 0.3740 | + | M SBV 2 62 -0.5728 0.3740 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 58 59 | + | M SAL 3 2 58 59 |
| − | M SBL 3 1 64 | + | M SBL 3 1 64 |
| − | M SMT 3 ^ CH2OH | + | M SMT 3 ^ CH2OH |
| − | M SBV 3 64 0.5584 -0.5305 | + | M SBV 3 64 0.5584 -0.5305 |
| − | S SKP 5 | + | S SKP 5 |
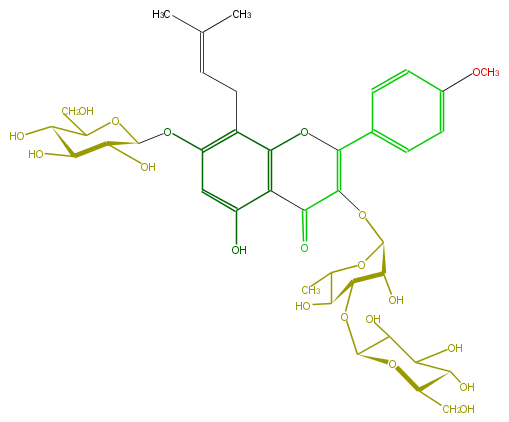
| − | ID FL5FABGI0012 | + | ID FL5FABGI0012 |
| − | FORMULA C39H50O20 | + | FORMULA C39H50O20 |
| − | EXACTMASS 838.28954404 | + | EXACTMASS 838.28954404 |
| − | AVERAGEMASS 838.8023000000001 | + | AVERAGEMASS 838.8023000000001 |
| − | SMILES O(c(c6)ccc(c6)C(O5)=C(C(c(c53)c(cc(OC(O4)C(O)C(O)C(O)C(CO)4)c3CC=C(C)C)O)=O)OC(O1)C(O)C(OC(C2O)OC(C(O)C2O)CO)C(C1C)O)C | + | SMILES O(c(c6)ccc(c6)C(O5)=C(C(c(c53)c(cc(OC(O4)C(O)C(O)C(O)C(CO)4)c3CC=C(C)C)O)=O)OC(O1)C(O)C(OC(C2O)OC(C(O)C2O)CO)C(C1C)O)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
59 64 0 0 0 0 0 0 0 0999 V2000
-1.4428 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4428 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7280 0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0132 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0132 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7280 2.1185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7015 0.4677 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4163 0.8804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4163 1.7058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7015 2.1185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7053 -0.3082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1308 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8593 1.6977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5878 2.1182 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5878 2.9594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8593 3.3801 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1308 2.9594 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1573 2.1182 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9228 0.3807 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7280 -0.3573 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7280 2.9435 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4425 3.3561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4425 4.1811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7280 4.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1570 4.5935 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5999 2.2262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1230 1.5966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4362 1.8637 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7736 1.8709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2552 2.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8560 2.0354 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2341 2.0231 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8387 1.6613 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6289 1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2659 -0.8352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2659 -1.4928 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7230 -1.0202 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3642 -0.8586 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3643 -0.2011 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9070 -0.6737 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5444 -1.7512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7614 -1.5181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7309 -1.1896 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5573 -1.3962 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4741 -2.1694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8047 -2.5559 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5546 -2.7436 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0959 -3.3014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7654 -2.9150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0155 -2.7273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0769 -1.8091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7945 -2.4019 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0396 -3.2254 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3161 3.3800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2341 2.8500 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6687 -3.6754 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1387 -4.5935 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4143 2.5659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7769 3.5619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
6 21 1 0 0 0 0
21 22 1 0 0 0 0
22 23 2 0 0 0 0
23 24 1 0 0 0 0
23 25 1 0 0 0 0
26 27 1 1 0 0 0
27 28 1 1 0 0 0
29 28 1 1 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 26 1 0 0 0 0
26 32 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 18 1 0 0 0 0
35 36 1 0 0 0 0
36 37 1 1 0 0 0
37 38 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 35 1 0 0 0 0
37 41 1 0 0 0 0
36 42 1 0 0 0 0
35 43 1 0 0 0 0
38 44 1 0 0 0 0
39 19 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 1 0 0 0
47 48 1 1 0 0 0
49 48 1 1 0 0 0
49 50 1 0 0 0 0
50 45 1 0 0 0 0
45 51 1 0 0 0 0
50 52 1 0 0 0 0
49 53 1 0 0 0 0
46 41 1 0 0 0 0
54 55 1 0 0 0 0
15 54 1 0 0 0 0
56 57 1 0 0 0 0
48 56 1 0 0 0 0
58 59 1 0 0 0 0
31 58 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 60
M SMT 1 OCH3
M SBV 1 60 -0.7283 -0.4205
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 56 57
M SBL 2 1 62
M SMT 2 CH2OH
M SBV 2 62 -0.5728 0.3740
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 58 59
M SBL 3 1 64
M SMT 3 ^ CH2OH
M SBV 3 64 0.5584 -0.5305
S SKP 5
ID FL5FABGI0012
FORMULA C39H50O20
EXACTMASS 838.28954404
AVERAGEMASS 838.8023000000001
SMILES O(c(c6)ccc(c6)C(O5)=C(C(c(c53)c(cc(OC(O4)C(O)C(O)C(O)C(CO)4)c3CC=C(C)C)O)=O)OC(O1)C(O)C(OC(C2O)OC(C(O)C2O)CO)C(C1C)O)C
M END