Mol:FL5FAAGS0055
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -3.8370 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8370 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8370 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8370 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2807 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2807 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7244 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7244 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7244 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7244 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2807 1.8981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2807 1.8981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1681 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1681 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6118 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6118 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6118 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6118 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1681 1.8981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1681 1.8981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1681 0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1681 0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9113 2.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9113 2.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3443 1.6944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3443 1.6944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2226 2.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2226 2.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2226 2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2226 2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3443 3.0037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3443 3.0037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9113 2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9113 2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2807 -0.0288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2807 -0.0288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0163 3.0557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0163 3.0557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3320 1.8981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3320 1.8981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0485 0.5250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0485 0.5250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2639 -0.5275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2639 -0.5275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3643 -0.8902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3643 -0.8902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1650 -0.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1650 -0.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3643 0.5090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3643 0.5090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2639 0.8718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2639 0.8718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0646 0.1742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0646 0.1742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7057 0.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7057 0.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1025 -1.1297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1025 -1.1297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3643 -1.5395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3643 -1.5395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7852 -0.5508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7852 -0.5508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6660 -2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6660 -2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3604 -2.5914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3604 -2.5914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2765 -2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2765 -2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5690 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5690 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2116 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2116 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4928 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4928 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0552 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0552 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3365 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3365 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0552 -2.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0552 -2.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4928 -2.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4928 -2.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8988 -2.5686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8988 -2.5686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7974 1.3273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7974 1.3273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0991 0.8048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0991 0.8048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7096 0.8048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7096 0.8048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0021 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0021 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6447 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6447 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9259 -0.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9259 -0.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4884 -0.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4884 -0.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7696 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7696 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4884 0.7853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4884 0.7853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9259 0.7853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9259 0.7853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3320 0.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3320 0.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 1 0 0 0 | + | 26 27 1 1 0 0 0 |
| − | 27 22 1 1 0 0 0 | + | 27 22 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 24 31 1 0 0 0 0 | + | 24 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 32 34 1 0 0 0 0 | + | 32 34 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 2 0 0 0 0 | + | 36 37 2 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 2 0 0 0 0 | + | 40 41 2 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 39 42 1 0 0 0 0 | + | 39 42 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 44 28 1 0 0 0 0 | + | 44 28 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0055 | + | ID FL5FAAGS0055 |
| − | KNApSAcK_ID C00005868 | + | KNApSAcK_ID C00005868 |
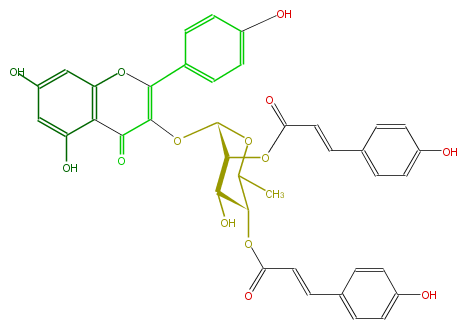
| − | NAME Kaempferol 3-(2'',4''-di-(E)-p-coumarylrhamnoside) | + | NAME Kaempferol 3-(2'',4''-di-(E)-p-coumarylrhamnoside) |
| − | CAS_RN 163434-73-9 | + | CAS_RN 163434-73-9 |
| − | FORMULA C39H32O14 | + | FORMULA C39H32O14 |
| − | EXACTMASS 724.179205732 | + | EXACTMASS 724.179205732 |
| − | AVERAGEMASS 724.6629800000001 | + | AVERAGEMASS 724.6629800000001 |
| − | SMILES c(c6)(ccc(c6)O)C=CC(OC(C(O)4)C(OC(C)C4OC(C=Cc(c5)ccc(O)c5)=O)OC(=C1c(c3)ccc(O)c3)C(=O)c(c2O)c(cc(c2)O)O1)=O | + | SMILES c(c6)(ccc(c6)O)C=CC(OC(C(O)4)C(OC(C)C4OC(C=Cc(c5)ccc(O)c5)=O)OC(=C1c(c3)ccc(O)c3)C(=O)c(c2O)c(cc(c2)O)O1)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-3.8370 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8370 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2807 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7244 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7244 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2807 1.8981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1681 0.6133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6118 0.9345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6118 1.5769 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1681 1.8981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1681 0.1125 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9113 2.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3443 1.6944 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2226 2.0217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2226 2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3443 3.0037 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9113 2.6764 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2807 -0.0288 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0163 3.0557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3320 1.8981 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0485 0.5250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2639 -0.5275 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3643 -0.8902 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1650 -0.1927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3643 0.5090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2639 0.8718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0646 0.1742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7057 0.1742 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1025 -1.1297 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3643 -1.5395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7852 -0.5508 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6660 -2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3604 -2.5914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2765 -2.0621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5690 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2116 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4928 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0552 -3.0557 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3365 -2.5686 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0552 -2.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4928 -2.0815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8988 -2.5686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7974 1.3273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0991 0.8048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7096 0.8048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0021 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6447 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9259 -0.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4884 -0.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7696 0.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4884 0.7853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9259 0.7853 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3320 0.2983 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 27 1 1 0 0 0
27 22 1 1 0 0 0
27 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
24 31 1 0 0 0 0
26 21 1 0 0 0 0
30 32 1 0 0 0 0
32 33 2 0 0 0 0
32 34 1 0 0 0 0
34 35 2 0 0 0 0
35 36 1 0 0 0 0
36 37 2 0 0 0 0
37 38 1 0 0 0 0
38 39 2 0 0 0 0
39 40 1 0 0 0 0
40 41 2 0 0 0 0
41 36 1 0 0 0 0
39 42 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
44 28 1 0 0 0 0
S SKP 8
ID FL5FAAGS0055
KNApSAcK_ID C00005868
NAME Kaempferol 3-(2'',4''-di-(E)-p-coumarylrhamnoside)
CAS_RN 163434-73-9
FORMULA C39H32O14
EXACTMASS 724.179205732
AVERAGEMASS 724.6629800000001
SMILES c(c6)(ccc(c6)O)C=CC(OC(C(O)4)C(OC(C)C4OC(C=Cc(c5)ccc(O)c5)=O)OC(=C1c(c3)ccc(O)c3)C(=O)c(c2O)c(cc(c2)O)O1)=O
M END