Mol:FL5FAAGS0028
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.3661 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3661 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3661 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3661 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1902 -0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1902 -0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7465 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7465 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7465 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7465 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1902 1.2582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1902 1.2582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3028 -0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3028 -0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8591 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8591 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8591 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8591 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3028 1.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3028 1.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3028 -0.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3028 -0.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4152 1.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4152 1.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9822 0.9308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9822 0.9308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5492 1.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5492 1.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5492 1.9128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5492 1.9128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9822 2.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9822 2.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4152 1.9128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4152 1.9128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1902 -0.6686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1902 -0.6686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9222 1.2581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9222 1.2581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1160 2.2400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1160 2.2400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5279 -0.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5279 -0.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5549 1.0750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5549 1.0750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0930 0.4654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0930 0.4654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4280 0.7240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4280 0.7240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7862 0.7310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7862 0.7310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2526 1.1974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2526 1.1974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8344 0.8903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8344 0.8903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1943 0.7059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1943 0.7059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8130 0.4304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8130 0.4304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0469 0.0843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0469 0.0843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1451 -1.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1451 -1.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7734 -1.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7734 -1.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5740 -0.9770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5740 -0.9770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7734 -0.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7734 -0.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1451 0.0876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1451 0.0876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3444 -0.6100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3444 -0.6100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1148 -0.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1148 -0.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3065 -1.9140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3065 -1.9140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6218 -2.2401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6218 -2.2401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1943 -1.3350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1943 -1.3350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9203 1.2887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9203 1.2887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7172 1.5023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7172 1.5023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 19 1 0 0 0 0 | + | 25 19 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 31 38 1 0 0 0 0 | + | 31 38 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 33 40 1 0 0 0 0 | + | 33 40 1 0 0 0 0 |
| − | 35 21 1 0 0 0 0 | + | 35 21 1 0 0 0 0 |
| − | 27 41 1 0 0 0 0 | + | 27 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 45 -6.4704 4.3261 | + | M SBV 1 45 -6.4704 4.3261 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGS0028 | + | ID FL5FAAGS0028 |
| − | KNApSAcK_ID C00005188 | + | KNApSAcK_ID C00005188 |
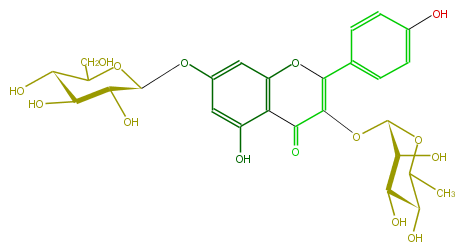
| − | NAME Kaempferol 3-rhamnoside-7-glucoside | + | NAME Kaempferol 3-rhamnoside-7-glucoside |
| − | CAS_RN 64323-49-5 | + | CAS_RN 64323-49-5 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(c(c42)C(=O)C(=C(c(c3)ccc(c3)O)O2)OC(C(O)1)OC(C)C(C1O)O)O | + | SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(c(c42)C(=O)C(=C(c(c3)ccc(c3)O)O2)OC(C(O)1)OC(C)C(C1O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-0.3661 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3661 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1902 -0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7465 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7465 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1902 1.2582 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3028 -0.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8591 0.2947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8591 0.9370 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3028 1.2582 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3028 -0.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4152 1.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9822 0.9308 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5492 1.2581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5492 1.9128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9822 2.2401 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4152 1.9128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1902 -0.6686 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9222 1.2581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1160 2.2400 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5279 -0.2143 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5549 1.0750 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0930 0.4654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4280 0.7240 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7862 0.7310 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2526 1.1974 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8344 0.8903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1943 0.7059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8130 0.4304 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0469 0.0843 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1451 -1.3117 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7734 -1.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5740 -0.9770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7734 -0.2752 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1451 0.0876 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3444 -0.6100 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1148 -0.6100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3065 -1.9140 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6218 -2.2401 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1943 -1.3350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9203 1.2887 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7172 1.5023 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 19 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
36 37 1 0 0 0 0
31 38 1 0 0 0 0
32 39 1 0 0 0 0
33 40 1 0 0 0 0
35 21 1 0 0 0 0
27 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 ^CH2OH
M SBV 1 45 -6.4704 4.3261
S SKP 8
ID FL5FAAGS0028
KNApSAcK_ID C00005188
NAME Kaempferol 3-rhamnoside-7-glucoside
CAS_RN 64323-49-5
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES C(C(O)5)(OC(CO)C(O)C5O)Oc(c4)cc(c(c42)C(=O)C(=C(c(c3)ccc(c3)O)O2)OC(C(O)1)OC(C)C(C1O)O)O
M END