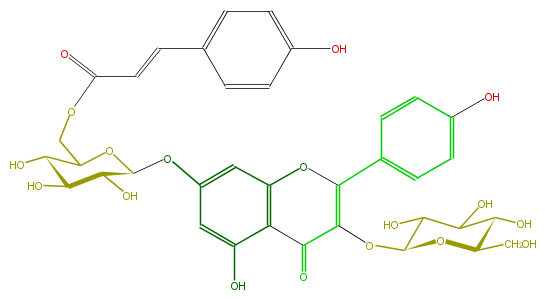
Mol:FL5FAAGL0087
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.4673 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4673 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4673 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4673 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7529 -1.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7529 -1.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0384 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0384 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0384 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0384 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7529 -0.2763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7529 -0.2763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6761 -1.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6761 -1.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3905 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3905 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3905 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3905 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6761 -0.2763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6761 -0.2763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6761 -2.5695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6761 -2.5695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2900 -0.1175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2900 -0.1175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0182 -0.5379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0182 -0.5379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7464 -0.1175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7464 -0.1175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7464 0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7464 0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0182 1.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0182 1.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2900 0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2900 0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7529 -2.7510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7529 -2.7510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5258 1.1733 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5258 1.1733 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1296 -0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1296 -0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0545 -1.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0545 -1.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1632 -1.3506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1632 -1.3506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7769 -2.0197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7769 -2.0197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5199 -1.8074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5199 -1.8074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2673 -2.0197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2673 -2.0197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6537 -1.3506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6537 -1.3506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9107 -1.5627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9107 -1.5627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5708 -1.4823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5708 -1.4823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3780 -1.0578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3780 -1.0578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1854 -1.4652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1854 -1.4652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6846 -0.0770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6846 -0.0770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2079 -0.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2079 -0.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5214 -0.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5214 -0.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8591 -0.4323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8591 -0.4323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3405 0.0492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3405 0.0492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9410 -0.2678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9410 -0.2678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1854 -0.2369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1854 -0.2369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8215 -0.6814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8215 -0.6814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9824 -0.8741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9824 -0.8741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3801 0.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3801 0.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1306 0.8673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1306 0.8673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6376 1.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6376 1.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7899 1.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7899 1.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2748 2.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2748 2.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3253 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3253 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3934 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3934 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9295 2.9408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9295 2.9408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0175 2.9408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0175 2.9408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4307 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4307 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0330 1.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0330 1.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9451 1.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9451 1.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3418 2.1581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3418 2.1581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9249 -1.8811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9249 -1.8811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9249 -2.9408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9249 -2.9408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 42 44 2 0 0 0 0 | + | 42 44 2 0 0 0 0 |
| − | 43 45 2 0 0 0 0 | + | 43 45 2 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 2 0 0 0 0 | + | 46 47 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 2 0 0 0 0 | + | 48 49 2 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 2 0 0 0 0 | + | 50 51 2 0 0 0 0 |
| − | 51 46 1 0 0 0 0 | + | 51 46 1 0 0 0 0 |
| − | 49 52 1 0 0 0 0 | + | 49 52 1 0 0 0 0 |
| − | 41 40 1 0 0 0 0 | + | 41 40 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 25 53 1 0 0 0 0 | + | 25 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 53 54 | + | M SAL 1 2 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 59 -0.6576 -0.1386 | + | M SBV 1 59 -0.6576 -0.1386 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0087 | + | ID FL5FAAGL0087 |
| − | FORMULA C36H36O18 | + | FORMULA C36H36O18 |
| − | EXACTMASS 756.190164348 | + | EXACTMASS 756.190164348 |
| − | AVERAGEMASS 756.6602399999999 | + | AVERAGEMASS 756.6602399999999 |
| − | SMILES OC(C5COC(=O)C=Cc(c6)ccc(c6)O)C(O)C(C(O5)Oc(c1)cc(O3)c(C(C(=C3c(c4)ccc(O)c4)OC(O2)C(C(C(O)C2CO)O)O)=O)c1O)O | + | SMILES OC(C5COC(=O)C=Cc(c6)ccc(c6)O)C(O)C(C(O5)Oc(c1)cc(O3)c(C(C(=C3c(c4)ccc(O)c4)OC(O2)C(C(C(O)C2CO)O)O)=O)c1O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.4673 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4673 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7529 -1.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0384 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0384 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7529 -0.2763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6761 -1.9262 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3905 -1.5138 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3905 -0.6888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6761 -0.2763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6761 -2.5695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2900 -0.1175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0182 -0.5379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7464 -0.1175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7464 0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0182 1.1438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2900 0.7233 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7529 -2.7510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5258 1.1733 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1296 -0.1060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0545 -1.9185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1632 -1.3506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7769 -2.0197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5199 -1.8074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2673 -2.0197 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6537 -1.3506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9107 -1.5627 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5708 -1.4823 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3780 -1.0578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1854 -1.4652 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6846 -0.0770 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2079 -0.7064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5214 -0.4394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8591 -0.4323 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3405 0.0492 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9410 -0.2678 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1854 -0.2369 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8215 -0.6814 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9824 -0.8741 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3801 0.2601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1306 0.8673 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6376 1.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7899 1.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2748 2.0668 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3253 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3934 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9295 2.9408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0175 2.9408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4307 2.1581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0330 1.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9451 1.3756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3418 2.1581 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9249 -1.8811 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9249 -2.9408 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
34 20 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
42 44 2 0 0 0 0
43 45 2 0 0 0 0
45 46 1 0 0 0 0
46 47 2 0 0 0 0
47 48 1 0 0 0 0
48 49 2 0 0 0 0
49 50 1 0 0 0 0
50 51 2 0 0 0 0
51 46 1 0 0 0 0
49 52 1 0 0 0 0
41 40 1 0 0 0 0
53 54 1 0 0 0 0
25 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 53 54
M SBL 1 1 59
M SMT 1 CH2OH
M SBV 1 59 -0.6576 -0.1386
S SKP 5
ID FL5FAAGL0087
FORMULA C36H36O18
EXACTMASS 756.190164348
AVERAGEMASS 756.6602399999999
SMILES OC(C5COC(=O)C=Cc(c6)ccc(c6)O)C(O)C(C(O5)Oc(c1)cc(O3)c(C(C(=C3c(c4)ccc(O)c4)OC(O2)C(C(C(O)C2CO)O)O)=O)c1O)O
M END