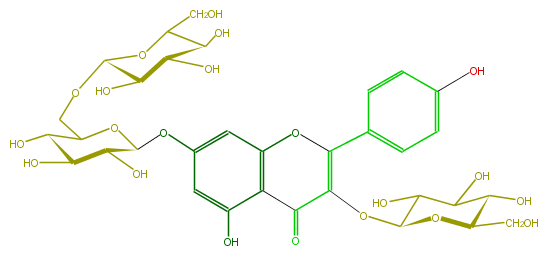
Mol:FL5FAAGL0052
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 54 59 0 0 0 0 0 0 0 0999 V2000 | + | 54 59 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6184 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6184 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6184 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6184 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9039 -2.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9039 -2.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1895 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1895 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1895 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1895 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9039 -0.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9039 -0.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5250 -2.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5250 -2.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2394 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2394 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2394 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2394 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5250 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5250 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5250 -2.6454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5250 -2.6454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0566 -0.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0566 -0.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7847 -0.7583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7847 -0.7583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5129 -0.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5129 -0.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5129 0.5030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5129 0.5030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7847 0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7847 0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0566 0.5030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0566 0.5030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9039 -2.6615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9039 -2.6615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2922 0.9529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2922 0.9529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2541 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2541 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9629 -2.1157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9629 -2.1157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1301 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1301 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7438 -2.3607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7438 -2.3607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4867 -2.1484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4867 -2.1484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2342 -2.3607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2342 -2.3607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6205 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6205 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8775 -1.9038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8775 -1.9038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3933 -1.8440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3933 -1.8440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3861 -1.2751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3861 -1.2751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2825 -1.7879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2825 -1.7879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7121 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7121 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1738 -1.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1738 -1.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4861 -0.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4861 -0.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8225 -0.7338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8225 -0.7338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3046 -0.2514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3046 -0.2514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0071 -0.5036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0071 -0.5036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2825 -0.5902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2825 -0.5902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9923 -0.9891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9923 -0.9891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8225 -1.2181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8225 -1.2181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4771 -0.0956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4771 -0.0956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5234 1.1597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5234 1.1597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7544 0.7188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7544 0.7188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2253 1.1999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2253 1.1999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3891 1.4587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3891 1.4587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1904 1.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1904 1.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7129 1.2770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7129 1.2770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1258 0.4801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1258 0.4801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4662 0.5360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4662 0.5360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3151 0.9959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3151 0.9959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0965 1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0965 1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6276 2.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6276 2.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1436 3.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1436 3.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0488 -2.2631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0488 -2.2631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0488 -3.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0488 -3.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 27 29 1 0 0 0 0 | + | 27 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 42 43 1 1 0 0 0 | + | 42 43 1 1 0 0 0 |
| − | 44 43 1 1 0 0 0 | + | 44 43 1 1 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 41 1 0 0 0 0 | + | 46 41 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 47 40 1 0 0 0 0 | + | 47 40 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 34 20 1 0 0 0 0 | + | 34 20 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 25 53 1 0 0 0 0 | + | 25 53 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 51 52 | + | M SAL 1 2 51 52 |
| − | M SBL 1 1 57 | + | M SBL 1 1 57 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 57 -0.5627 -0.3810 | + | M SBV 1 57 -0.5627 -0.3810 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 53 54 | + | M SAL 2 2 53 54 |
| − | M SBL 2 1 59 | + | M SBL 2 1 59 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 59 -0.8146 -0.0976 | + | M SBV 2 59 -0.8146 -0.0976 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGL0052 | + | ID FL5FAAGL0052 |
| − | FORMULA C33H40O21 | + | FORMULA C33H40O21 |
| − | EXACTMASS 772.206208342 | + | EXACTMASS 772.206208342 |
| − | AVERAGEMASS 772.6581 | + | AVERAGEMASS 772.6581 |
| − | SMILES O=C(C(OC(C6O)OC(CO)C(C6O)O)=4)c(c(O)3)c(OC4c(c5)ccc(O)c5)cc(c3)OC(O1)C(C(O)C(C1COC(C2O)OC(CO)C(O)C2O)O)O | + | SMILES O=C(C(OC(C6O)OC(CO)C(C6O)O)=4)c(c(O)3)c(OC4c(c5)ccc(O)c5)cc(c3)OC(O1)C(C(O)C(C1COC(C2O)OC(CO)C(O)C2O)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
54 59 0 0 0 0 0 0 0 0999 V2000
-1.6184 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6184 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9039 -2.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1895 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1895 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9039 -0.3522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5250 -2.0022 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2394 -1.5897 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2394 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5250 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5250 -2.6454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0566 -0.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7847 -0.7583 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5129 -0.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5129 0.5030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7847 0.9234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0566 0.5030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9039 -2.6615 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2922 0.9529 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2541 -0.3522 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9629 -2.1157 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1301 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7438 -2.3607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4867 -2.1484 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2342 -2.3607 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6205 -1.6916 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8775 -1.9038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3933 -1.8440 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3861 -1.2751 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2825 -1.7879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7121 -0.4157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1738 -1.0083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4861 -0.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8225 -0.7338 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3046 -0.2514 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0071 -0.5036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2825 -0.5902 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9923 -0.9891 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8225 -1.2181 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4771 -0.0956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5234 1.1597 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7544 0.7188 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2253 1.1999 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3891 1.4587 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1904 1.7654 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7129 1.2770 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1258 0.4801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4662 0.5360 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3151 0.9959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0965 1.7533 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6276 2.1465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1436 3.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0488 -2.2631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0488 -3.3227 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
4 3 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
27 29 1 0 0 0 0
26 30 1 0 0 0 0
23 21 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
36 40 1 0 0 0 0
41 42 1 1 0 0 0
42 43 1 1 0 0 0
44 43 1 1 0 0 0
44 45 1 0 0 0 0
45 46 1 0 0 0 0
46 41 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 49 1 0 0 0 0
47 40 1 0 0 0 0
44 50 1 0 0 0 0
34 20 1 0 0 0 0
51 52 1 0 0 0 0
45 51 1 0 0 0 0
53 54 1 0 0 0 0
25 53 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 51 52
M SBL 1 1 57
M SMT 1 CH2OH
M SBV 1 57 -0.5627 -0.3810
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 53 54
M SBL 2 1 59
M SMT 2 CH2OH
M SBV 2 59 -0.8146 -0.0976
S SKP 5
ID FL5FAAGL0052
FORMULA C33H40O21
EXACTMASS 772.206208342
AVERAGEMASS 772.6581
SMILES O=C(C(OC(C6O)OC(CO)C(C6O)O)=4)c(c(O)3)c(OC4c(c5)ccc(O)c5)cc(c3)OC(O1)C(C(O)C(C1COC(C2O)OC(CO)C(O)C2O)O)O
M END