Mol:FL5FAAGI0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 48 52 0 0 0 0 0 0 0 0999 V2000 | + | 48 52 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3407 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3407 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3407 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3407 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7844 -1.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7844 -1.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2281 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2281 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2281 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2281 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7844 0.0070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7844 0.0070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3282 -1.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3282 -1.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8845 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8845 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8845 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8845 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3282 0.0070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3282 0.0070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3282 -1.7786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3282 -1.7786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4406 0.0069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4406 0.0069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0076 -0.3205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0076 -0.3205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5746 0.0069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5746 0.0069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5746 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5746 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0076 0.9889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0076 0.9889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4406 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4406 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8968 0.0069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8968 0.0069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1414 0.9888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1414 0.9888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2787 -1.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2787 -1.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7844 -1.9199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7844 -1.9199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7844 0.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7844 0.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3405 0.9702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3405 0.9702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3405 1.6123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3405 1.6123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7844 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7844 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8966 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8966 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8300 -0.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8300 -0.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4588 -0.5596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4588 -0.5596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9244 -0.3518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9244 -0.3518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4086 -0.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4086 -0.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7834 0.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7834 0.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2510 -0.2181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2510 -0.2181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3439 -0.3663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3439 -0.3663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0375 -0.5878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0375 -0.5878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6181 -0.8659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6181 -0.8659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4168 -1.0266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4168 -1.0266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1160 -1.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1160 -1.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6945 -1.3822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6945 -1.3822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2764 -1.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2764 -1.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5773 -1.0266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5773 -1.0266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9988 -1.1918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9988 -1.1918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8581 -1.1763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8581 -1.1763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2370 -0.7793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2370 -0.7793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0114 -1.2772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0114 -1.2772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6294 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6294 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3439 -1.9334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3439 -1.9334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6494 0.5660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6494 0.5660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8525 0.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8525 0.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 6 22 1 0 0 0 0 | + | 6 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 18 1 0 0 0 0 | + | 30 18 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | 37 20 1 0 0 0 0 | + | 37 20 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 32 47 1 0 0 0 0 | + | 32 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 45 46 | + | M SAL 1 2 45 46 |
| − | M SBL 1 1 49 | + | M SBL 1 1 49 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 49 -5.6036 3.5694 | + | M SBV 1 49 -5.6036 3.5694 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 47 48 | + | M SAL 2 2 47 48 |
| − | M SBL 2 1 51 | + | M SBL 2 1 51 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 51 -6.3550 4.3269 | + | M SBV 2 51 -6.3550 4.3269 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FAAGI0007 | + | ID FL5FAAGI0007 |
| − | KNApSAcK_ID C00005810 | + | KNApSAcK_ID C00005810 |
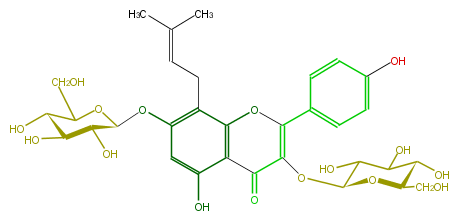
| − | NAME Hexandraside E;8-Prenylkaempferol 3,7-diglucoside;Noranhydroicaritin 3,7-diglucoside;3,4',5,7-Tetrahydroxy-8-(3-methyl-2-butenyl)flavone 3,7-diglucoside;3,7-Bis(beta-D-glucopyranosyloxy)-5-hydroxy-2-(4-hydroxyphenyl)-8-(3-methyl-2-butenyl)-4H-1-benzopyran-4-one | + | NAME Hexandraside E;8-Prenylkaempferol 3,7-diglucoside;Noranhydroicaritin 3,7-diglucoside;3,4',5,7-Tetrahydroxy-8-(3-methyl-2-butenyl)flavone 3,7-diglucoside;3,7-Bis(beta-D-glucopyranosyloxy)-5-hydroxy-2-(4-hydroxyphenyl)-8-(3-methyl-2-butenyl)-4H-1-benzopyran-4-one |
| − | CAS_RN 139955-75-2 | + | CAS_RN 139955-75-2 |
| − | FORMULA C32H38O16 | + | FORMULA C32H38O16 |
| − | EXACTMASS 678.215985168 | + | EXACTMASS 678.215985168 |
| − | AVERAGEMASS 678.63452 | + | AVERAGEMASS 678.63452 |
| − | SMILES c(c(O)4)(C2=O)c(c(CC=C(C)C)c(OC(C5O)OC(C(O)C5O)CO)c4)OC(=C(OC(C3O)OC(C(C3O)O)CO)2)c(c1)ccc(c1)O | + | SMILES c(c(O)4)(C2=O)c(c(CC=C(C)C)c(OC(C5O)OC(C(O)C5O)CO)c4)OC(=C(OC(C3O)OC(C(C3O)O)CO)2)c(c1)ccc(c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
48 52 0 0 0 0 0 0 0 0999 V2000
-1.3407 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3407 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7844 -1.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2281 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2281 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7844 0.0070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3282 -1.2777 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8845 -0.9565 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8845 -0.3142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3282 0.0070 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3282 -1.7786 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4406 0.0069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0076 -0.3205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5746 0.0069 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5746 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0076 0.9889 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4406 0.6616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8968 0.0069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1414 0.9888 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2787 -1.3454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7844 -1.9199 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7844 0.6491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3405 0.9702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3405 1.6123 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7844 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8966 1.9334 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8300 -0.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4588 -0.5596 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9244 -0.3518 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4086 -0.3462 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7834 0.0287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2510 -0.2181 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3439 -0.3663 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0375 -0.5878 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6181 -0.8659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4168 -1.0266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1160 -1.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6945 -1.3822 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2764 -1.5476 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5773 -1.0266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9988 -1.1918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8581 -1.1763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2370 -0.7793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0114 -1.2772 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6294 -1.5209 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3439 -1.9334 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6494 0.5660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8525 0.3525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
6 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
24 26 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 18 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 1 0 0 0
38 39 1 1 0 0 0
40 39 1 1 0 0 0
40 41 1 0 0 0 0
41 36 1 0 0 0 0
36 42 1 0 0 0 0
41 43 1 0 0 0 0
40 44 1 0 0 0 0
37 20 1 0 0 0 0
39 45 1 0 0 0 0
45 46 1 0 0 0 0
32 47 1 0 0 0 0
47 48 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 45 46
M SBL 1 1 49
M SMT 1 CH2OH
M SBV 1 49 -5.6036 3.5694
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 47 48
M SBL 2 1 51
M SMT 2 ^CH2OH
M SBV 2 51 -6.3550 4.3269
S SKP 8
ID FL5FAAGI0007
KNApSAcK_ID C00005810
NAME Hexandraside E;8-Prenylkaempferol 3,7-diglucoside;Noranhydroicaritin 3,7-diglucoside;3,4',5,7-Tetrahydroxy-8-(3-methyl-2-butenyl)flavone 3,7-diglucoside;3,7-Bis(beta-D-glucopyranosyloxy)-5-hydroxy-2-(4-hydroxyphenyl)-8-(3-methyl-2-butenyl)-4H-1-benzopyran-4-one
CAS_RN 139955-75-2
FORMULA C32H38O16
EXACTMASS 678.215985168
AVERAGEMASS 678.63452
SMILES c(c(O)4)(C2=O)c(c(CC=C(C)C)c(OC(C5O)OC(C(O)C5O)CO)c4)OC(=C(OC(C3O)OC(C(C3O)O)CO)2)c(c1)ccc(c1)O
M END