Mol:FL3FECGS0021
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.9295 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9295 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9295 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9295 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3805 -2.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3805 -2.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8316 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8316 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8316 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8316 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3805 -1.6724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3805 -1.6724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2826 -2.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2826 -2.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7337 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7337 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7337 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7337 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2826 -1.6724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2826 -1.6724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2826 -3.1202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2826 -3.1202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1846 -1.6725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1846 -1.6725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6443 -1.9379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6443 -1.9379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1041 -1.6725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1041 -1.6725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1041 -1.1417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1041 -1.1417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6443 -0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6443 -0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1846 -1.1417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1846 -1.1417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6443 -0.2725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6443 -0.2725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6943 -0.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6943 -0.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3805 -3.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3805 -3.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4793 -1.6730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4793 -1.6730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4793 -2.7136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4793 -2.7136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1834 0.3460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1834 0.3460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6691 -0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6691 -0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3289 -0.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3289 -0.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3289 -1.4231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3289 -1.4231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1568 -0.9375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1568 -0.9375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1834 -0.3407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1834 -0.3407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4048 0.7296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4048 0.7296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6691 0.3676 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6691 0.3676 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7052 -1.1127 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7052 -1.1127 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5007 -1.3040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5007 -1.3040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9851 -1.9846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9851 -1.9846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2426 -1.6958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2426 -1.6958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5261 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5261 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0468 -1.1674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0468 -1.1674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6963 -1.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6963 -1.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2145 -1.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2145 -1.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7889 -2.0237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7889 -2.0237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8172 -2.4100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8172 -2.4100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2140 -0.9926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2140 -0.9926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8505 -0.3631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8505 -0.3631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4988 0.0111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4988 0.0111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0611 -0.3135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0611 -0.3135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4988 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4988 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1273 0.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1273 0.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1273 1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1273 1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5604 1.8307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5604 1.8307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5604 2.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5604 2.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1273 2.8127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1273 2.8127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6943 2.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6943 2.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6943 1.8307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6943 1.8307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1273 3.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1273 3.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2274 0.2211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2274 0.2211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9419 -0.1914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9419 -0.1914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 16 18 1 0 0 0 0 | + | 16 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 1 21 1 0 0 0 0 | + | 1 21 1 0 0 0 0 |
| − | 2 22 1 0 0 0 0 | + | 2 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 31 1 0 0 0 0 | + | 35 31 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 28 54 1 0 0 0 0 | + | 28 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 59 -5.6639 4.5378 | + | M SBV 1 59 -5.6639 4.5378 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FECGS0021 | + | ID FL3FECGS0021 |
| − | KNApSAcK_ID C00004396 | + | KNApSAcK_ID C00004396 |
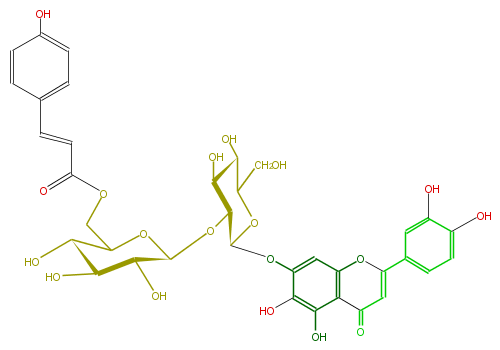
| − | NAME 6-Hydroxyluteoin-7-(6'''-p-coumarylsophoroside) | + | NAME 6-Hydroxyluteoin-7-(6'''-p-coumarylsophoroside) |
| − | CAS_RN 113720-08-4 | + | CAS_RN 113720-08-4 |
| − | FORMULA C36H36O19 | + | FORMULA C36H36O19 |
| − | EXACTMASS 772.18507897 | + | EXACTMASS 772.18507897 |
| − | AVERAGEMASS 772.6596400000001 | + | AVERAGEMASS 772.6596400000001 |
| − | SMILES C(=C5)(c(c6)ccc(c6O)O)Oc(c1)c(C5=O)c(O)c(O)c(OC(O2)C(OC(C4O)OC(C(O)C4O)COC(=O)C=Cc(c3)ccc(c3)O)C(C(O)C(CO)2)O)1 | + | SMILES C(=C5)(c(c6)ccc(c6O)O)Oc(c1)c(C5=O)c(O)c(O)c(OC(O2)C(OC(C4O)OC(C(O)C4O)COC(=O)C=Cc(c3)ccc(c3)O)C(C(O)C(CO)2)O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
0.9295 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9295 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3805 -2.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8316 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8316 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3805 -1.6724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2826 -2.7141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7337 -2.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7337 -1.9328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2826 -1.6724 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2826 -3.1202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1846 -1.6725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6443 -1.9379 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1041 -1.6725 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1041 -1.1417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6443 -0.8763 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1846 -1.1417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6443 -0.2725 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6943 -0.8009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3805 -3.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4793 -1.6730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4793 -2.7136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1834 0.3460 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6691 -0.1396 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3289 -0.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3289 -1.4231 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1568 -0.9375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1834 -0.3407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4048 0.7296 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6691 0.3676 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7052 -1.1127 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5007 -1.3040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9851 -1.9846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2426 -1.6958 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5261 -1.6881 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0468 -1.1674 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6963 -1.5102 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2145 -1.7161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7889 -2.0237 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8172 -2.4100 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2140 -0.9926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8505 -0.3631 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4988 0.0111 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0611 -0.3135 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4988 0.6358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1273 0.8042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1273 1.5034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5604 1.8307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5604 2.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1273 2.8127 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6943 2.4854 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6943 1.8307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1273 3.2339 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2274 0.2211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9419 -0.1914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
16 18 1 0 0 0 0
19 15 1 0 0 0 0
3 20 1 0 0 0 0
1 21 1 0 0 0 0
2 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 21 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 31 1 0 0 0 0
37 41 1 0 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
28 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 59
M SMT 1 CH2OH
M SBV 1 59 -5.6639 4.5378
S SKP 8
ID FL3FECGS0021
KNApSAcK_ID C00004396
NAME 6-Hydroxyluteoin-7-(6'''-p-coumarylsophoroside)
CAS_RN 113720-08-4
FORMULA C36H36O19
EXACTMASS 772.18507897
AVERAGEMASS 772.6596400000001
SMILES C(=C5)(c(c6)ccc(c6O)O)Oc(c1)c(C5=O)c(O)c(O)c(OC(O2)C(OC(C4O)OC(C(O)C4O)COC(=O)C=Cc(c3)ccc(c3)O)C(C(O)C(CO)2)O)1
M END