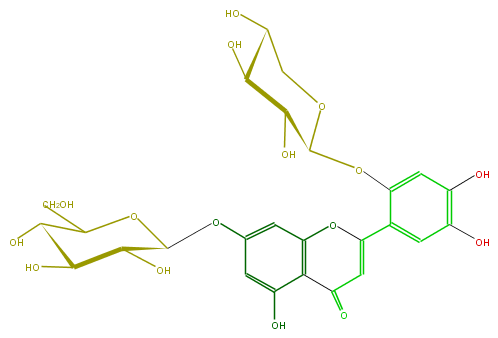
Mol:FL3FALGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.4097 -1.6942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4097 -1.6942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4097 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4097 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1085 -2.9684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1085 -2.9684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8072 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8072 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8072 -1.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8072 -1.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1085 -1.3547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1085 -1.3547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5060 -2.9684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5060 -2.9684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2048 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2048 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2048 -1.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2048 -1.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5060 -1.3547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5060 -1.3547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7719 -3.5383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7719 -3.5383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9033 -1.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9033 -1.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6154 -1.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6154 -1.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3275 -1.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3275 -1.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3275 -0.5324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3275 -0.5324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6154 -0.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6154 -0.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9033 -0.5324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9033 -0.5324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1085 -3.7735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1085 -3.7735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0395 -0.1214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0395 -0.1214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0395 -1.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0395 -1.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3048 -1.2817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3048 -1.2817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1513 -0.0197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1513 -0.0197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9258 3.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9258 3.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4121 2.1434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4121 2.1434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3746 1.3710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3746 1.3710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9399 0.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9399 0.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2353 1.5176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2353 1.5176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2824 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2824 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1785 3.7735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1785 3.7735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0478 3.0046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0478 3.0046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3454 0.3694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3454 0.3694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5464 -1.3417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5464 -1.3417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7477 -2.3960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7477 -2.3960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5974 -1.9488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5974 -1.9488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4877 -1.9367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4877 -1.9367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2941 -1.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2941 -1.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4690 -1.5519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4690 -1.5519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2112 -1.7555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2112 -1.7555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6615 -2.3754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6615 -2.3754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6676 -2.4391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6676 -2.4391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5593 -0.8416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5593 -0.8416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0395 -0.3847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0395 -0.3847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 21 1 1 0 0 0 0 | + | 21 1 1 0 0 0 0 |
| − | 17 22 1 0 0 0 0 | + | 17 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 25 31 1 0 0 0 0 | + | 25 31 1 0 0 0 0 |
| − | 26 22 1 0 0 0 0 | + | 26 22 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 21 1 0 0 0 0 | + | 35 21 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 37 41 1 0 0 0 0 | + | 37 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 46 1.0903 -0.7102 | + | M SBV 1 46 1.0903 -0.7102 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FALGS0004 | + | ID FL3FALGS0004 |
| − | FORMULA C26H28O16 | + | FORMULA C26H28O16 |
| − | EXACTMASS 596.137734848 | + | EXACTMASS 596.137734848 |
| − | AVERAGEMASS 596.49092 | + | AVERAGEMASS 596.49092 |
| − | SMILES c(c(C(=C3)Oc(c4)c(c(cc(OC(C(O)5)OC(CO)C(O)C5O)4)O)C3=O)2)(cc(O)c(O)c2)OC(C(O)1)OCC(O)C1O | + | SMILES c(c(C(=C3)Oc(c4)c(c(cc(OC(C(O)5)OC(CO)C(O)C5O)4)O)C3=O)2)(cc(O)c(O)c2)OC(C(O)1)OCC(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
0.4097 -1.6942 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4097 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1085 -2.9684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8072 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8072 -1.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1085 -1.3547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5060 -2.9684 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2048 -2.5650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2048 -1.7581 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5060 -1.3547 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7719 -3.5383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9033 -1.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6154 -1.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3275 -1.3548 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3275 -0.5324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6154 -0.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9033 -0.5324 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1085 -3.7735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0395 -0.1214 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.0395 -1.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3048 -1.2817 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1513 -0.0197 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9258 3.3624 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4121 2.1434 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3746 1.3710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9399 0.4158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2353 1.5176 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2824 2.3242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1785 3.7735 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0478 3.0046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3454 0.3694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5464 -1.3417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7477 -2.3960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5974 -1.9488 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4877 -1.9367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2941 -1.1301 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4690 -1.5519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2112 -1.7555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6615 -2.3754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6676 -2.4391 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5593 -0.8416 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0395 -0.3847 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
15 19 1 0 0 0 0
14 20 1 0 0 0 0
21 1 1 0 0 0 0
17 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
25 31 1 0 0 0 0
26 22 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 21 1 0 0 0 0
41 42 1 0 0 0 0
37 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 ^ CH2OH
M SBV 1 46 1.0903 -0.7102
S SKP 5
ID FL3FALGS0004
FORMULA C26H28O16
EXACTMASS 596.137734848
AVERAGEMASS 596.49092
SMILES c(c(C(=C3)Oc(c4)c(c(cc(OC(C(O)5)OC(CO)C(O)C5O)4)O)C3=O)2)(cc(O)c(O)c2)OC(C(O)1)OCC(O)C1O
M END