Mol:FL3FACDS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2595 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2595 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2595 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2595 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5509 -2.2538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5509 -2.2538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1577 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1577 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1577 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1577 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5509 -0.6174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5509 -0.6174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8663 -2.2538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8663 -2.2538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5749 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5749 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5749 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5749 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8663 -0.6174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8663 -0.6174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8663 -2.8918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8663 -2.8918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5509 -3.0718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5509 -3.0718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4410 -0.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4410 -0.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1877 -0.9173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1877 -0.9173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9344 -0.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9344 -0.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9344 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9344 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1877 0.8071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1877 0.8071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4410 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4410 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6807 0.8069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6807 0.8069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0582 -1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0582 -1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4013 -2.4175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4013 -2.4175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7446 -2.0234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7446 -2.0234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8514 -2.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8514 -2.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4819 -1.5504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4819 -1.5504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1912 -1.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1912 -1.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6807 -2.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6807 -2.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0053 -2.5793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0053 -2.5793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8861 -0.5007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8861 -0.5007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9241 -2.4971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9241 -2.4971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5314 2.4169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5314 2.4169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3214 1.8266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3214 1.8266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9860 1.1380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9860 1.1380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1686 0.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1686 0.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5379 0.8351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5379 0.8351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8162 1.5715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8162 1.5715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9096 3.0718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9096 3.0718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3548 2.6510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3548 2.6510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5688 0.5553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5688 0.5553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1877 1.6688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1877 1.6688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7634 -1.3198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7634 -1.3198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3693 -0.1994 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3693 -0.1994 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 17 39 1 0 0 0 0 | + | 17 39 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.5722 -0.5722 | + | M SBV 1 45 0.5722 -0.5722 |
| − | S SKP 5 | + | S SKP 5 |
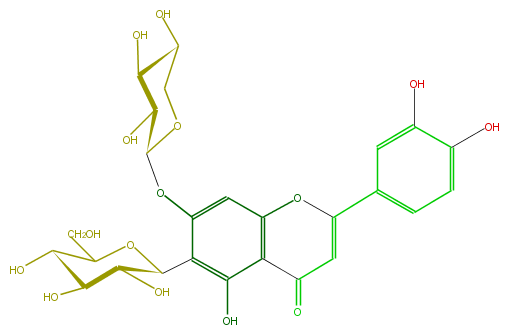
| − | ID FL3FACDS0001 | + | ID FL3FACDS0001 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES O(c(c5)c(c(O)c(c54)C(=O)C=C(O4)c(c3)ccc(O)c3O)C(C(O)2)OC(C(O)C(O)2)CO)C(C(O)1)OCC(O)C1O | + | SMILES O(c(c5)c(c(O)c(c54)C(=O)C=C(O4)c(c3)ccc(O)c3O)C(C(O)2)OC(C(O)C(O)2)CO)C(C(O)1)OCC(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-1.2595 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2595 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5509 -2.2538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1577 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1577 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5509 -0.6174 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8663 -2.2538 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5749 -1.8448 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5749 -1.0265 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8663 -0.6174 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8663 -2.8918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5509 -3.0718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4410 -0.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1877 -0.9173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9344 -0.4862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9344 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1877 0.8071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4410 0.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6807 0.8069 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0582 -1.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4013 -2.4175 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7446 -2.0234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8514 -2.1284 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4819 -1.5504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1912 -1.8920 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.6807 -2.0412 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0053 -2.5793 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8861 -0.5007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9241 -2.4971 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5314 2.4169 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3214 1.8266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9860 1.1380 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1686 0.2573 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5379 0.8351 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8162 1.5715 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9096 3.0718 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3548 2.6510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5688 0.5553 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1877 1.6688 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7634 -1.3198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3693 -0.1994 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 28 1 0 0 0 0
17 39 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.5722 -0.5722
S SKP 5
ID FL3FACDS0001
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES O(c(c5)c(c(O)c(c54)C(=O)C=C(O4)c(c3)ccc(O)c3O)C(C(O)2)OC(C(O)C(O)2)CO)C(C(O)1)OCC(O)C1O
M END