Mol:FL3FACCS0026
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0751 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0751 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0751 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0751 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3606 -2.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3606 -2.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6461 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6461 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6461 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6461 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3606 -0.4658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3606 -0.4658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0683 -2.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0683 -2.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7828 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7828 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7828 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7828 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0683 -0.4658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0683 -0.4658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0683 -2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0683 -2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7893 -0.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7893 -0.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5765 -0.4395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5765 -0.4395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3293 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3293 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0822 -0.4395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0822 -0.4395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0822 0.4299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0822 0.4299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3293 0.8645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3293 0.8645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5765 0.4299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5765 0.4299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8347 0.8642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8347 0.8642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2235 2.1420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2235 2.1420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1559 1.8543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1559 1.8543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1360 0.9442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1360 0.9442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4077 0.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4077 0.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6451 0.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6451 0.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7593 1.3589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7593 1.3589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2553 2.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2553 2.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8205 2.6282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8205 2.6282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8822 0.4763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8822 0.4763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8347 -0.8739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8347 -0.8739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3606 -2.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3606 -2.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1878 1.2071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1878 1.2071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6872 0.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6872 0.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6321 1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6321 1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1734 2.0221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1734 2.0221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6740 2.6189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6740 2.6189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7291 1.8420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7291 1.8420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1224 2.8761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1224 2.8761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0072 2.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0072 2.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1185 2.4277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1185 2.4277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8347 1.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8347 1.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0879 1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0879 1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5932 0.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5932 0.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 13 9 1 0 0 0 0 | + | 13 9 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 6 23 1 0 0 0 0 | + | 6 23 1 0 0 0 0 |
| − | 15 29 1 0 0 0 0 | + | 15 29 1 0 0 0 0 |
| − | 3 30 1 0 0 0 0 | + | 3 30 1 0 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 36 31 1 1 0 0 0 | + | 36 31 1 1 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 34 38 1 0 0 0 0 | + | 34 38 1 0 0 0 0 |
| − | 36 39 1 0 0 0 0 | + | 36 39 1 0 0 0 0 |
| − | 31 40 1 0 0 0 0 | + | 31 40 1 0 0 0 0 |
| − | 32 28 1 0 0 0 0 | + | 32 28 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 25 41 1 0 0 0 0 | + | 25 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 46 | + | M SBL 1 1 46 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 46 -0.6714 -0.2963 | + | M SBV 1 46 -0.6714 -0.2963 |
| − | S SKP 5 | + | S SKP 5 |
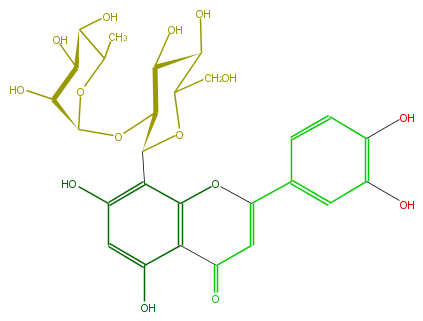
| − | ID FL3FACCS0026 | + | ID FL3FACCS0026 |
| − | FORMULA C27H30O15 | + | FORMULA C27H30O15 |
| − | EXACTMASS 594.15847029 | + | EXACTMASS 594.15847029 |
| − | AVERAGEMASS 594.5181 | + | AVERAGEMASS 594.5181 |
| − | SMILES OC(C2O)C(C(c(c34)c(cc(c3C(C=C(c(c5)ccc(O)c5O)O4)=O)O)O)OC2CO)OC(O1)C(O)C(O)C(C1C)O | + | SMILES OC(C2O)C(C(c(c34)c(cc(c3C(C=C(c(c5)ccc(O)c5O)O4)=O)O)O)OC2CO)OC(O1)C(O)C(O)C(C1C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.0751 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0751 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3606 -2.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6461 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6461 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3606 -0.4658 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0683 -2.1157 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7828 -1.7034 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7828 -0.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0683 -0.4658 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0683 -2.7591 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7893 -0.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5765 -0.4395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3293 -0.8742 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0822 -0.4395 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0822 0.4299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3293 0.8645 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5765 0.4299 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8347 0.8642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2235 2.1420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1559 1.8543 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1360 0.9442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4077 0.1718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6451 0.5273 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7593 1.3589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2553 2.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8205 2.6282 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8822 0.4763 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8347 -0.8739 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3606 -2.9405 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1878 1.2071 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6872 0.6104 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6321 1.3874 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1734 2.0221 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6740 2.6189 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7291 1.8420 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1224 2.8761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0072 2.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1185 2.4277 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8347 1.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0879 1.6552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5932 0.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
13 9 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
6 23 1 0 0 0 0
15 29 1 0 0 0 0
3 30 1 0 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 36 1 1 0 0 0
36 31 1 1 0 0 0
35 37 1 0 0 0 0
34 38 1 0 0 0 0
36 39 1 0 0 0 0
31 40 1 0 0 0 0
32 28 1 0 0 0 0
41 42 1 0 0 0 0
25 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 46
M SMT 1 CH2OH
M SBV 1 46 -0.6714 -0.2963
S SKP 5
ID FL3FACCS0026
FORMULA C27H30O15
EXACTMASS 594.15847029
AVERAGEMASS 594.5181
SMILES OC(C2O)C(C(c(c34)c(cc(c3C(C=C(c(c5)ccc(O)c5O)O4)=O)O)O)OC2CO)OC(O1)C(O)C(O)C(C1C)O
M END