Mol:FL3FAADS0019
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.6593 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6593 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6593 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6593 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1030 -0.7926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1030 -0.7926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5467 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5467 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5467 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5467 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1030 0.4922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1030 0.4922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0096 -0.7926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0096 -0.7926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5659 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5659 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5659 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5659 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0096 0.4922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0096 0.4922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0096 -1.2934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0096 -1.2934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2154 0.4920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2154 0.4920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1030 -1.4347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1030 -1.4347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2458 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2458 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8320 0.2567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8320 0.2567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4182 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4182 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4182 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4182 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8320 1.6105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8320 1.6105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2458 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2458 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0041 1.6103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0041 1.6103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2468 4.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2468 4.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6412 3.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6412 3.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8981 3.0885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8981 3.0885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9050 2.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9050 2.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3683 2.9142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3683 2.9142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0633 3.4922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0633 3.4922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7788 5.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7788 5.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6064 4.4643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6064 4.4643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2627 2.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2627 2.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8392 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8392 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4680 -1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4680 -1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9336 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9336 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4178 -0.9748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4178 -0.9748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7926 -0.5999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7926 -0.5999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2602 -0.8467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2602 -0.8467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3531 -0.9949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3531 -0.9949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0467 -1.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0467 -1.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6273 -1.4945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6273 -1.4945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6273 -2.1626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6273 -2.1626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2766 -2.3366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2766 -2.3366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1439 -2.4417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1439 -2.4417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1439 -3.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1439 -3.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4269 -3.4797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4269 -3.4797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4269 -4.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4269 -4.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8693 -4.4453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8693 -4.4453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3118 -4.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3118 -4.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3118 -3.4797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3118 -3.4797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8693 -3.1578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8693 -3.1578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2448 -4.4448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2448 -4.4448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6586 -0.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6586 -0.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8617 -0.2761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8617 -0.2761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3055 -4.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3055 -4.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5910 -5.0185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5910 -5.0185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6386 3.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6386 3.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3531 3.4118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3531 3.4118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 1 12 1 0 0 0 0 | + | 1 12 1 0 0 0 0 |
| − | 3 13 1 0 0 0 0 | + | 3 13 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 9 14 1 0 0 0 0 | + | 9 14 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 20 24 1 0 0 0 0 | + | 20 24 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 2 33 1 0 0 0 0 | + | 2 33 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 35 50 1 0 0 0 0 | + | 35 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 45 52 1 0 0 0 0 | + | 45 52 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 26 54 1 0 0 0 0 | + | 26 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 ^CH2OH | + | M SMT 1 ^CH2OH |
| − | M SBV 1 55 -7.1067 7.5552 | + | M SBV 1 55 -7.1067 7.5552 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 57 -7.1444 6.6103 | + | M SBV 2 57 -7.1444 6.6103 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 54 55 | + | M SAL 3 2 54 55 |
| − | M SBL 3 1 59 | + | M SBL 3 1 59 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SBV 3 59 -6.1330 7.1031 | + | M SBV 3 59 -6.1330 7.1031 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FAADS0019 | + | ID FL3FAADS0019 |
| − | KNApSAcK_ID C00006332 | + | KNApSAcK_ID C00006332 |
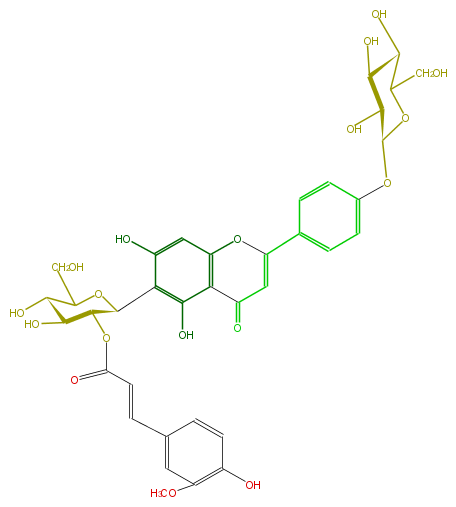
| − | NAME Isovitexin 4'-O-glucoside 2''-O-(E)-ferulate | + | NAME Isovitexin 4'-O-glucoside 2''-O-(E)-ferulate |
| − | CAS_RN 64763-02-6 | + | CAS_RN 64763-02-6 |
| − | FORMULA C37H38O18 | + | FORMULA C37H38O18 |
| − | EXACTMASS 770.205814412 | + | EXACTMASS 770.205814412 |
| − | AVERAGEMASS 770.6868199999999 | + | AVERAGEMASS 770.6868199999999 |
| − | SMILES C(OC(C6O)C(OC(C6O)CO)c(c(O)2)c(O)cc(O3)c2C(C=C3c(c4)ccc(OC(C(O)5)OC(CO)C(C5O)O)c4)=O)(=O)C=Cc(c1)cc(OC)c(O)c1 | + | SMILES C(OC(C6O)C(OC(C6O)CO)c(c(O)2)c(O)cc(O3)c2C(C=C3c(c4)ccc(OC(C(O)5)OC(CO)C(C5O)O)c4)=O)(=O)C=Cc(c1)cc(OC)c(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.6593 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6593 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1030 -0.7926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5467 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5467 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1030 0.4922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0096 -0.7926 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5659 -0.4714 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5659 0.1710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0096 0.4922 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0096 -1.2934 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.2154 0.4920 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1030 -1.4347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2458 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8320 0.2567 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4182 0.5952 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4182 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8320 1.6105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2458 1.2721 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0041 1.6103 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2468 4.2079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6412 3.7491 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8981 3.0885 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9050 2.4510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.3683 2.9142 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0633 3.4922 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.7788 5.0185 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6064 4.4643 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2627 2.7099 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.8392 -0.6983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4680 -1.1882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9336 -0.9803 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4178 -0.9748 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7926 -0.5999 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2602 -0.8467 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3531 -0.9949 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0467 -1.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6273 -1.4945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6273 -2.1626 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2766 -2.3366 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1439 -2.4417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1439 -3.1291 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4269 -3.4797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4269 -4.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8693 -4.4453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3118 -4.1234 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3118 -3.4797 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8693 -3.1578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2448 -4.4448 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6586 -0.0625 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8617 -0.2761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3055 -4.6060 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5910 -5.0185 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6386 3.8243 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3531 3.4118 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
1 12 1 0 0 0 0
3 13 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
9 14 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
20 24 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
2 33 1 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
39 41 1 0 0 0 0
41 42 2 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 43 1 0 0 0 0
46 49 1 0 0 0 0
35 50 1 0 0 0 0
50 51 1 0 0 0 0
45 52 1 0 0 0 0
52 53 1 0 0 0 0
26 54 1 0 0 0 0
54 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 55
M SMT 1 ^CH2OH
M SBV 1 55 -7.1067 7.5552
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 57
M SMT 2 OCH3
M SBV 2 57 -7.1444 6.6103
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 54 55
M SBL 3 1 59
M SMT 3 CH2OH
M SBV 3 59 -6.1330 7.1031
S SKP 8
ID FL3FAADS0019
KNApSAcK_ID C00006332
NAME Isovitexin 4'-O-glucoside 2''-O-(E)-ferulate
CAS_RN 64763-02-6
FORMULA C37H38O18
EXACTMASS 770.205814412
AVERAGEMASS 770.6868199999999
SMILES C(OC(C6O)C(OC(C6O)CO)c(c(O)2)c(O)cc(O3)c2C(C=C3c(c4)ccc(OC(C(O)5)OC(CO)C(C5O)O)c4)=O)(=O)C=Cc(c1)cc(OC)c(O)c1
M END