Mol:FL2FADGS0004
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 51 56 0 0 0 0 0 0 0 0999 V2000 | + | 51 56 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0169 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0169 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7321 -1.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7321 -1.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4474 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4474 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4474 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4474 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7321 -0.1590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7321 -0.1590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0169 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0169 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1625 -1.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1625 -1.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8777 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8777 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8777 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8777 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1625 -0.1590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1625 -0.1590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1625 -2.5300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1625 -2.5300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7321 -2.6357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7321 -2.6357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6788 -0.1703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6788 -0.1703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6736 -0.1608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6736 -0.1608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3996 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3996 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1257 -0.1608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1257 -0.1608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1257 0.6775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1257 0.6775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3996 1.0967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3996 1.0967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6736 0.6775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6736 0.6775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8200 1.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8200 1.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1174 -0.3091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1174 -0.3091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6449 -0.9329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6449 -0.9329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9645 -0.6683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9645 -0.6683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3081 -0.6612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3081 -0.6612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7850 -0.1841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7850 -0.1841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4801 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4801 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7095 -0.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7095 -0.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3666 -0.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3666 -0.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4233 -1.1645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4233 -1.1645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1231 -1.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1231 -1.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7007 -2.3105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7007 -2.3105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8052 -2.2929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8052 -2.2929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4264 -1.9106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4264 -1.9106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2760 -1.4645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2760 -1.4645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7311 -1.8724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7311 -1.8724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6993 -3.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6993 -3.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8052 -2.9347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8052 -2.9347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2758 -1.9815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2758 -1.9815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8326 -2.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8326 -2.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9896 -2.4345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9896 -2.4345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5572 -1.9603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5572 -1.9603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4272 -1.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4272 -1.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8312 -3.0798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8312 -3.0798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9977 -3.1287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9977 -3.1287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5785 -1.3198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5785 -1.3198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8239 -2.0193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8239 -2.0193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8200 -1.7576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8200 -1.7576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3996 1.8850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3996 1.8850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9816 3.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9816 3.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0134 0.0665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0134 0.0665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1370 1.1235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1370 1.1235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 2 12 1 0 0 0 0 | + | 2 12 1 0 0 0 0 |
| − | 6 13 1 0 0 0 0 | + | 6 13 1 0 0 0 0 |
| − | 9 14 1 6 0 0 0 | + | 9 14 1 6 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 13 1 0 0 0 0 | + | 24 13 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 31 36 1 0 0 0 0 | + | 31 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 39 40 1 1 0 0 0 | + | 39 40 1 1 0 0 0 |
| − | 41 40 1 1 0 0 0 | + | 41 40 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 38 1 0 0 0 0 | + | 42 38 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | 41 45 1 0 0 0 0 | + | 41 45 1 0 0 0 0 |
| − | 45 35 1 0 0 0 0 | + | 45 35 1 0 0 0 0 |
| − | 33 29 1 0 0 0 0 | + | 33 29 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 39 46 1 0 0 0 0 | + | 39 46 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 18 48 1 0 0 0 0 | + | 18 48 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 26 50 1 0 0 0 0 | + | 26 50 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 46 47 | + | M SAL 1 2 46 47 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 52 -0.0088 -0.4220 | + | M SBV 1 52 -0.0088 -0.4220 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 48 49 | + | M SAL 2 2 48 49 |
| − | M SBL 2 1 54 | + | M SBL 2 1 54 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 54 0.0000 -0.7883 | + | M SBV 2 54 0.0000 -0.7883 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 50 51 | + | M SAL 3 2 50 51 |
| − | M SBL 3 1 56 | + | M SBL 3 1 56 |
| − | M SMT 3 ^ CH2OH | + | M SMT 3 ^ CH2OH |
| − | M SBV 3 56 0.5333 -0.5000 | + | M SBV 3 56 0.5333 -0.5000 |
| − | S SKP 5 | + | S SKP 5 |
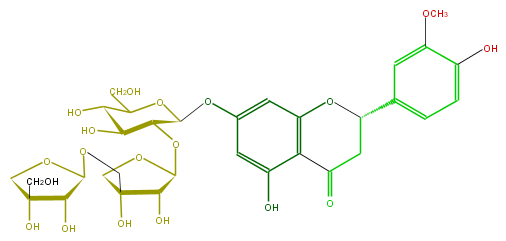
| − | ID FL2FADGS0004 | + | ID FL2FADGS0004 |
| − | FORMULA C32H40O19 | + | FORMULA C32H40O19 |
| − | EXACTMASS 728.216379098 | + | EXACTMASS 728.216379098 |
| − | AVERAGEMASS 728.6486 | + | AVERAGEMASS 728.6486 |
| − | SMILES C(O)C(C(O)1)OC(Oc(c6)cc(c(c56)C(CC(O5)c(c4)cc(c(c4)O)OC)=O)O)C(OC(O2)C(O)C(COC(C(O)3)OCC(O)3CO)(C2)O)C1O | + | SMILES C(O)C(C(O)1)OC(Oc(c6)cc(c(c56)C(CC(O5)c(c4)cc(c(c4)O)OC)=O)O)C(OC(O2)C(O)C(COC(C(O)3)OCC(O)3CO)(C2)O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
51 56 0 0 0 0 0 0 0 0999 V2000
0.0169 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7321 -1.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4474 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4474 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7321 -0.1590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0169 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1625 -1.8107 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8777 -1.3977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8777 -0.5718 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1625 -0.1590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1625 -2.5300 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7321 -2.6357 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6788 -0.1703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6736 -0.1608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3996 -0.5798 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1257 -0.1608 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1257 0.6775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3996 1.0967 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6736 0.6775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8200 1.0783 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1174 -0.3091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6449 -0.9329 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9645 -0.6683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3081 -0.6612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7850 -0.1841 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4801 -0.4336 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.7095 -0.4202 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3666 -0.8375 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4233 -1.1645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1231 -1.9126 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7007 -2.3105 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8052 -2.2929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4264 -1.9106 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2760 -1.4645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7311 -1.8724 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6993 -3.0110 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8052 -2.9347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2758 -1.9815 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8326 -2.4413 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9896 -2.4345 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5572 -1.9603 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4272 -1.5555 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8312 -3.0798 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9977 -3.1287 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5785 -1.3198 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8239 -2.0193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8200 -1.7576 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3996 1.8850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.9816 3.1287 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0134 0.0665 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1370 1.1235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
3 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 4 1 0 0 0 0
7 11 2 0 0 0 0
2 12 1 0 0 0 0
6 13 1 0 0 0 0
9 14 1 6 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 13 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 30 1 0 0 0 0
31 35 1 0 0 0 0
31 36 1 0 0 0 0
32 37 1 0 0 0 0
38 39 1 1 0 0 0
39 40 1 1 0 0 0
41 40 1 1 0 0 0
41 42 1 0 0 0 0
42 38 1 0 0 0 0
39 43 1 0 0 0 0
40 44 1 0 0 0 0
41 45 1 0 0 0 0
45 35 1 0 0 0 0
33 29 1 0 0 0 0
46 47 1 0 0 0 0
39 46 1 0 0 0 0
48 49 1 0 0 0 0
18 48 1 0 0 0 0
50 51 1 0 0 0 0
26 50 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 46 47
M SBL 1 1 52
M SMT 1 CH2OH
M SBV 1 52 -0.0088 -0.4220
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 48 49
M SBL 2 1 54
M SMT 2 OCH3
M SBV 2 54 0.0000 -0.7883
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 50 51
M SBL 3 1 56
M SMT 3 ^ CH2OH
M SBV 3 56 0.5333 -0.5000
S SKP 5
ID FL2FADGS0004
FORMULA C32H40O19
EXACTMASS 728.216379098
AVERAGEMASS 728.6486
SMILES C(O)C(C(O)1)OC(Oc(c6)cc(c(c56)C(CC(O5)c(c4)cc(c(c4)O)OC)=O)O)C(OC(O2)C(O)C(COC(C(O)3)OCC(O)3CO)(C2)O)C1O
M END