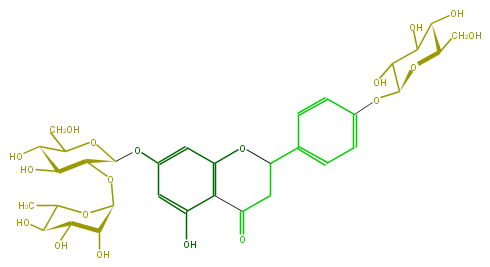
Mol:FL2FAAGS0020
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 52 57 0 0 0 0 0 0 0 0999 V2000 | + | 52 57 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3630 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3630 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3630 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3630 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5814 -2.1968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5814 -2.1968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7999 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7999 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7999 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7999 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5814 -0.3918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5814 -0.3918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0183 -2.1968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0183 -2.1968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7632 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7632 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7632 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7632 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0183 -0.3918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0183 -0.3918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5444 -0.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5444 -0.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3410 -0.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3410 -0.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1375 -0.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1375 -0.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1375 0.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1375 0.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3410 0.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3410 0.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5444 0.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5444 0.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0183 -2.9607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0183 -2.9607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7355 0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7355 0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9701 -0.4620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9701 -0.4620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5814 -3.0989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5814 -3.0989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1840 1.9593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1840 1.9593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3963 1.1668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3963 1.1668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7948 1.8842 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7948 1.8842 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5154 2.2859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5154 2.2859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3032 3.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3032 3.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9047 2.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9047 2.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7471 1.4495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7471 1.4495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7614 3.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7614 3.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9106 3.3815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9106 3.3815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.7202 -0.3621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.7202 -0.3621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1799 -1.0756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1799 -1.0756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4015 -0.7729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4015 -0.7729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5902 -0.7818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5902 -0.7818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1962 -0.2189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1962 -0.2189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9913 -0.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9913 -0.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.2782 -0.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.2782 -0.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9695 -1.0002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9695 -1.0002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7079 -1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7079 -1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1994 -2.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1994 -2.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.6058 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.6058 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8244 -2.5133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8244 -2.5133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0383 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0383 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6318 -2.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6318 -2.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4134 -2.2559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4134 -2.2559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9231 -2.0028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9231 -2.0028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0676 -2.4788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0676 -2.4788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2071 -3.1215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2071 -3.1215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0378 -3.3815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0378 -3.3815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9366 2.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9366 2.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.0165 1.6343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.0165 1.6343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4778 0.1367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4778 0.1367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -7.0165 0.3985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -7.0165 0.3985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 18 1 0 0 0 0 | + | 22 18 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 30 1 0 0 0 0 | + | 35 30 1 0 0 0 0 |
| − | 30 36 1 0 0 0 0 | + | 30 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 1 0 0 0 | + | 40 41 1 1 0 0 0 |
| − | 41 42 1 1 0 0 0 | + | 41 42 1 1 0 0 0 |
| − | 43 42 1 1 0 0 0 | + | 43 42 1 1 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 39 1 0 0 0 0 | + | 44 39 1 0 0 0 0 |
| − | 39 45 1 0 0 0 0 | + | 39 45 1 0 0 0 0 |
| − | 40 46 1 0 0 0 0 | + | 40 46 1 0 0 0 0 |
| − | 41 47 1 0 0 0 0 | + | 41 47 1 0 0 0 0 |
| − | 42 48 1 0 0 0 0 | + | 42 48 1 0 0 0 0 |
| − | 43 38 1 0 0 0 0 | + | 43 38 1 0 0 0 0 |
| − | 33 19 1 0 0 0 0 | + | 33 19 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 24 49 1 0 0 0 0 | + | 24 49 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 35 51 1 0 0 0 0 | + | 35 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 49 50 | + | M SAL 1 2 49 50 |
| − | M SBL 1 1 55 | + | M SBL 1 1 55 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 55 -0.4213 -0.4753 | + | M SBV 1 55 -0.4213 -0.4753 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 51 52 | + | M SAL 2 2 51 52 |
| − | M SBL 2 1 57 | + | M SBL 2 1 57 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 57 0.4865 -0.6411 | + | M SBV 2 57 0.4865 -0.6411 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL2FAAGS0020 | + | ID FL2FAAGS0020 |
| − | FORMULA C33H42O19 | + | FORMULA C33H42O19 |
| − | EXACTMASS 742.2320291619999 | + | EXACTMASS 742.2320291619999 |
| − | AVERAGEMASS 742.67518 | + | AVERAGEMASS 742.67518 |
| − | SMILES C(C(O1)C(O)C(C(O)C(Oc(c2)ccc(C(C3)Oc(c4)c(c(cc4OC(C(OC(O6)C(C(C(O)C6C)O)O)5)OC(CO)C(C(O)5)O)O)C(=O)3)c2)1)O)O | + | SMILES C(C(O1)C(O)C(C(O)C(Oc(c2)ccc(C(C3)Oc(c4)c(c(cc4OC(C(OC(O6)C(C(C(O)C6C)O)O)5)OC(CO)C(C(O)5)O)O)C(=O)3)c2)1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
52 57 0 0 0 0 0 0 0 0999 V2000
-2.3630 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3630 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5814 -2.1968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7999 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7999 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5814 -0.3918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0183 -2.1968 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7632 -1.7455 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7632 -0.8430 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0183 -0.3918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5444 -0.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3410 -0.8519 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1375 -0.3919 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1375 0.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3410 0.9878 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5444 0.5277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0183 -2.9607 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7355 0.9570 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9701 -0.4620 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5814 -3.0989 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1840 1.9593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3963 1.1668 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7948 1.8842 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5154 2.2859 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3032 3.0785 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9047 2.3613 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7471 1.4495 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.7614 3.0074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9106 3.3815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.7202 -0.3621 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1799 -1.0756 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4015 -0.7729 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5902 -0.7818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1962 -0.2189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.9913 -0.5045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.2782 -0.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9695 -1.0002 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7079 -1.2767 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1994 -2.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.6058 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8244 -2.5133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0383 -2.7364 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6318 -2.0327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.4134 -2.2559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.9231 -2.0028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.0676 -2.4788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2071 -3.1215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0378 -3.3815 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.9366 2.7612 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.0165 1.6343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4778 0.1367 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-7.0165 0.3985 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
14 18 1 0 0 0 0
1 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 18 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 35 1 0 0 0 0
35 30 1 0 0 0 0
30 36 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 1 0 0 0
41 42 1 1 0 0 0
43 42 1 1 0 0 0
43 44 1 0 0 0 0
44 39 1 0 0 0 0
39 45 1 0 0 0 0
40 46 1 0 0 0 0
41 47 1 0 0 0 0
42 48 1 0 0 0 0
43 38 1 0 0 0 0
33 19 1 0 0 0 0
49 50 1 0 0 0 0
24 49 1 0 0 0 0
51 52 1 0 0 0 0
35 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 49 50
M SBL 1 1 55
M SMT 1 CH2OH
M SBV 1 55 -0.4213 -0.4753
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 51 52
M SBL 2 1 57
M SMT 2 ^ CH2OH
M SBV 2 57 0.4865 -0.6411
S SKP 5
ID FL2FAAGS0020
FORMULA C33H42O19
EXACTMASS 742.2320291619999
AVERAGEMASS 742.67518
SMILES C(C(O1)C(O)C(C(O)C(Oc(c2)ccc(C(C3)Oc(c4)c(c(cc4OC(C(OC(O6)C(C(C(O)C6C)O)O)5)OC(CO)C(C(O)5)O)O)C(=O)3)c2)1)O)O
M END