Mol:FL1A3CGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0911 -1.2056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0911 -1.2056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0911 -0.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0911 -0.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4281 -0.3063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4281 -0.3063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9473 -0.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9473 -0.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9473 -1.2056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9473 -1.2056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4281 -1.5054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4281 -1.5054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6613 -1.3909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6613 -1.3909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0137 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0137 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6613 -0.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6613 -0.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4665 -0.3063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4665 -0.3063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4555 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4555 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3393 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3393 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6396 -1.4259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6396 -1.4259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2402 -1.4259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2402 -1.4259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5404 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5404 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2402 -0.3857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2402 -0.3857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6396 -0.3857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6396 -0.3857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1399 -0.9058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1399 -0.9058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8056 -1.9295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8056 -1.9295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5399 0.1335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5399 0.1335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6261 -0.3217 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.6261 -0.3217 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.2549 -0.8117 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.2549 -0.8117 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.7204 -0.6038 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.7204 -0.6038 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2047 -0.5982 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.2047 -0.5982 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5795 -0.2234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5795 -0.2234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0471 -0.4702 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.0471 -0.4702 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.9663 0.1642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9663 0.1642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6054 -1.2729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6054 -1.2729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4142 -1.1179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4142 -1.1179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0185 -0.2327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0185 -0.2327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4281 0.2932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4281 0.2932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4328 0.5710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4328 0.5710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9901 1.0137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9901 1.0137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2542 1.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2542 1.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5241 1.0137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5241 1.0137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2630 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2630 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6969 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6969 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4292 1.0023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4292 1.0023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8939 1.0023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8939 1.0023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6263 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6263 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8939 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8939 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4292 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4292 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0914 1.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0914 1.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 1 7 1 0 0 0 0 | + | 1 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 2 1 0 0 0 0 | + | 9 2 1 0 0 0 0 |
| − | 4 10 1 0 0 0 0 | + | 4 10 1 0 0 0 0 |
| − | 8 11 2 0 0 0 0 | + | 8 11 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 15 18 1 0 0 0 0 | + | 15 18 1 0 0 0 0 |
| − | 7 19 2 0 0 0 0 | + | 7 19 2 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 24 10 1 0 0 0 0 | + | 24 10 1 0 0 0 0 |
| − | 3 31 1 0 0 0 0 | + | 3 31 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL1A3CGS0003 | + | ID FL1A3CGS0003 |
| − | KNApSAcK_ID C00008052 | + | KNApSAcK_ID C00008052 |
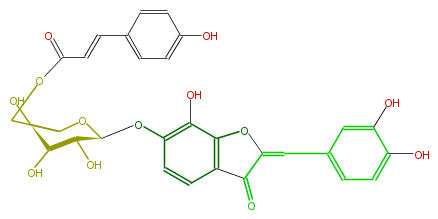
| − | NAME Maritimetin 6-(6''-p-coumarylglucoside) | + | NAME Maritimetin 6-(6''-p-coumarylglucoside) |
| − | CAS_RN 134955-55-8 | + | CAS_RN 134955-55-8 |
| − | FORMULA C30H26O13 | + | FORMULA C30H26O13 |
| − | EXACTMASS 594.137340918 | + | EXACTMASS 594.137340918 |
| − | AVERAGEMASS 594.51964 | + | AVERAGEMASS 594.51964 |
| − | SMILES Oc(c1)c(ccc1C=c(c(=O)2)oc(c3O)c2ccc3O[C@@H]([C@H]5O)OC([C@H](O)[C@@H]5O)COC(C=Cc(c4)ccc(O)c4)=O)O | + | SMILES Oc(c1)c(ccc1C=c(c(=O)2)oc(c3O)c2ccc3O[C@@H]([C@H]5O)OC([C@H](O)[C@@H]5O)COC(C=Cc(c4)ccc(O)c4)=O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
0.0911 -1.2056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0911 -0.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4281 -0.3063 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9473 -0.6060 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9473 -1.2056 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4281 -1.5054 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6613 -1.3909 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0137 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6613 -0.4208 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4665 -0.3063 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4555 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3393 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6396 -1.4259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2402 -1.4259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5404 -0.9058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2402 -0.3857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6396 -0.3857 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1399 -0.9058 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8056 -1.9295 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5399 0.1335 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6261 -0.3217 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.2549 -0.8117 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.7204 -0.6038 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2047 -0.5982 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5795 -0.2234 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0471 -0.4702 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.9663 0.1642 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.6054 -1.2729 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4142 -1.1179 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0185 -0.2327 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4281 0.2932 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4328 0.5710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9901 1.0137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2542 1.4710 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.5241 1.0137 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2630 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6969 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4292 1.0023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8939 1.0023 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6263 1.4659 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8939 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4292 1.9295 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0914 1.4659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
1 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 2 1 0 0 0 0
4 10 1 0 0 0 0
8 11 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
15 18 1 0 0 0 0
7 19 2 0 0 0 0
16 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
26 30 1 0 0 0 0
24 10 1 0 0 0 0
3 31 1 0 0 0 0
30 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
40 43 1 0 0 0 0
S SKP 8
ID FL1A3CGS0003
KNApSAcK_ID C00008052
NAME Maritimetin 6-(6''-p-coumarylglucoside)
CAS_RN 134955-55-8
FORMULA C30H26O13
EXACTMASS 594.137340918
AVERAGEMASS 594.51964
SMILES Oc(c1)c(ccc1C=c(c(=O)2)oc(c3O)c2ccc3O[C@@H]([C@H]5O)OC([C@H](O)[C@@H]5O)COC(C=Cc(c4)ccc(O)c4)=O)O
M END