Mol:BMFYB5CAa008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 58 60 0 0 1 0 0 0 0 0999 V2000 | + | 58 60 0 0 1 0 0 0 0 0999 V2000 |
| − | 2.6691 -4.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6691 -4.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6473 -4.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6473 -4.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3164 -3.9388 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 4.3164 -3.9388 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 4.9855 -3.1956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9855 -3.1956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9637 -3.4035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9637 -3.4035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0596 -4.6079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0596 -4.6079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 -5.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 -5.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3601 -3.5230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3601 -3.5230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5733 -3.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5733 -3.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2727 -4.3546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2727 -4.3546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.3208 -2.3736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.3208 -2.3736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.4548 -1.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.4548 -1.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.4548 -0.8736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.4548 -0.8736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.3208 -0.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.3208 -0.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.1868 -0.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.1868 -0.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.1868 -1.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.1868 -1.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.9300 -0.2045 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.9300 -0.2045 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.5232 0.7091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.5232 0.7091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.5287 0.6045 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.5287 0.6045 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 25.0528 -2.3736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 25.0528 -2.3736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.8596 1.3477 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 22.8596 1.3477 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 23.0675 2.3258 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 23.0675 2.3258 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 22.2015 2.8258 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 22.2015 2.8258 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 21.4583 2.1567 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 21.4583 2.1567 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 20.4802 2.3646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.4802 2.3646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.9810 2.7326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.9810 2.7326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.0969 3.8204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.0969 3.8204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.8651 1.2432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.8651 1.2432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.8110 1.6215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.8110 1.6215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.9060 4.4081 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.9060 4.4081 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.4937 3.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.4937 3.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.7150 4.9959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.7150 4.9959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.3182 5.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.3182 5.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.8329 1.8294 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.8329 1.8294 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.6250 0.8512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.6250 0.8512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.0408 2.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.0408 2.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.8547 2.0373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.8547 2.0373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.1856 1.2941 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.1856 1.2941 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.9288 0.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.9288 0.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.4425 1.9633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.4425 1.9633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.5165 0.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.5165 0.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.5383 0.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.5383 0.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.8692 0.0158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.8692 0.0158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.6124 -0.6534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.6124 -0.6534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.1261 0.6849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.1261 0.6849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.2001 -0.7274 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 14.2001 -0.7274 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 13.2219 -0.5195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.2219 -0.5195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.5528 -1.2626 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.5528 -1.2626 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.5747 -1.0547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.5747 -1.0547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.9055 -1.7978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.9055 -1.7978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.9274 -1.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.9274 -1.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.2582 -2.3331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.2582 -2.3331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.2801 -2.1252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.2801 -2.1252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.6110 -2.8683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.6110 -2.8683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.6328 -2.6604 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.6328 -2.6604 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.5091 -1.6784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.5091 -1.6784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.9129 0.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.9129 0.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.6184 -0.6389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.6184 -0.6389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11 16 2 0 0 0 0 | + | 11 16 2 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 15 14 2 0 0 0 0 | + | 15 14 2 0 0 0 0 |
| − | 19 18 1 0 0 0 0 | + | 19 18 1 0 0 0 0 |
| − | 18 17 2 0 0 0 0 | + | 18 17 2 0 0 0 0 |
| − | 17 15 1 0 0 0 0 | + | 17 15 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 24 28 1 6 0 0 0 | + | 24 28 1 6 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 21 28 1 6 0 0 0 | + | 21 28 1 6 0 0 0 |
| − | 23 22 1 0 0 0 0 | + | 23 22 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 29 34 1 0 0 0 0 | + | 29 34 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 41 38 1 0 0 0 0 | + | 41 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 38 37 1 0 0 0 0 | + | 38 37 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 46 1 0 0 0 0 | + | 43 46 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 46 56 1 1 0 0 0 | + | 46 56 1 1 0 0 0 |
| − | 47 57 2 0 0 0 0 | + | 47 57 2 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 51 58 2 0 0 0 0 | + | 51 58 2 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 22 26 1 1 0 0 0 | + | 22 26 1 1 0 0 0 |
| − | 23 27 1 1 0 0 0 | + | 23 27 1 1 0 0 0 |
| − | 31 30 1 0 0 0 0 | + | 31 30 1 0 0 0 0 |
| − | 30 32 1 0 0 0 0 | + | 30 32 1 0 0 0 0 |
| − | 30 33 2 0 0 0 0 | + | 30 33 2 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 55 5 1 0 0 0 0 | + | 55 5 1 0 0 0 0 |
| − | 5 4 1 0 0 0 0 | + | 5 4 1 0 0 0 0 |
| − | 5 10 2 0 0 0 0 | + | 5 10 2 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 3 9 1 6 0 0 0 | + | 3 9 1 6 0 0 0 |
| − | 3 6 1 1 0 0 0 | + | 3 6 1 1 0 0 0 |
| − | 1 8 1 0 0 0 0 | + | 1 8 1 0 0 0 0 |
| − | 1 7 2 0 0 0 0 | + | 1 7 2 0 0 0 0 |
| − | 2 1 1 0 0 0 0 | + | 2 1 1 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMFYB5CAa008 | + | ID BMFYB5CAa008 |
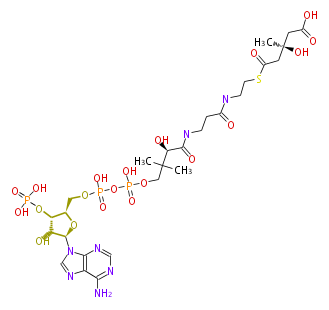
| − | NAME (S)-3-Hydroxy-3-methyl-glutaryl-CoA | + | NAME (S)-3-Hydroxy-3-methyl-glutaryl-CoA |
| − | FORMULA C27H44N7O20P3S | + | FORMULA C27H44N7O20P3S |
| − | EXACTMASS 911.1574 | + | EXACTMASS 911.1574 |
| − | AVERAGEMASS 911.6607 | + | AVERAGEMASS 911.6607 |
| − | SMILES C([C@H](C(NCCC(NCCSC(C[C@@](C)(O)CC(O)=O)=O)=O)=O)O)(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)3)O[C@H]([C@@H]3O)n(c21)cnc(c(N)ncn2)1)(O)=O)(O)=O | + | SMILES C([C@H](C(NCCC(NCCSC(C[C@@](C)(O)CC(O)=O)=O)=O)=O)O)(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)3)O[C@H]([C@@H]3O)n(c21)cnc(c(N)ncn2)1)(O)=O)(O)=O |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00356 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00356 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
58 60 0 0 1 0 0 0 0 0999 V2000
2.6691 -4.4740 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6473 -4.6819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3164 -3.9388 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
4.9855 -3.1956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9637 -3.4035 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0596 -4.6079 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 -5.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3601 -3.5230 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5733 -3.2696 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
6.2727 -4.3546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
23.3208 -2.3736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.4548 -1.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
22.4548 -0.8736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
23.3208 -0.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.1868 -0.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.1868 -1.8736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.9300 -0.2045 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
24.5232 0.7091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
23.5287 0.6045 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
25.0528 -2.3736 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.8596 1.3477 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
23.0675 2.3258 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
22.2015 2.8258 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
21.4583 2.1567 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
20.4802 2.3646 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
23.9810 2.7326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.0969 3.8204 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21.8651 1.2432 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.8110 1.6215 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.9060 4.4081 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
23.4937 3.5991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
23.7150 4.9959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.3182 5.2172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
18.8329 1.8294 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
18.6250 0.8512 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.0408 2.8075 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17.8547 2.0373 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17.1856 1.2941 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
17.9288 0.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.4425 1.9633 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.5165 0.5510 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.5383 0.7589 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.8692 0.0158 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
15.6124 -0.6534 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.1261 0.6849 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.2001 -0.7274 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
13.2219 -0.5195 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.5528 -1.2626 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
11.5747 -1.0547 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.9055 -1.7978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.9274 -1.5899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.2582 -2.3331 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
8.2801 -2.1252 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.6110 -2.8683 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.6328 -2.6604 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
14.5091 -1.6784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.9129 0.4316 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.6184 -0.6389 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
11 16 2 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
15 16 1 0 0 0 0
15 14 2 0 0 0 0
19 18 1 0 0 0 0
18 17 2 0 0 0 0
17 15 1 0 0 0 0
14 19 1 0 0 0 0
16 20 1 0 0 0 0
24 28 1 6 0 0 0
23 24 1 0 0 0 0
21 28 1 6 0 0 0
23 22 1 0 0 0 0
21 22 1 0 0 0 0
24 25 1 0 0 0 0
25 29 1 0 0 0 0
21 19 1 0 0 0 0
29 34 1 0 0 0 0
34 36 1 0 0 0 0
34 35 2 0 0 0 0
34 37 1 0 0 0 0
41 38 1 0 0 0 0
38 39 2 0 0 0 0
38 40 1 0 0 0 0
41 42 1 0 0 0 0
38 37 1 0 0 0 0
42 43 1 0 0 0 0
43 46 1 0 0 0 0
43 44 1 0 0 0 0
43 45 1 0 0 0 0
46 47 1 0 0 0 0
46 56 1 1 0 0 0
47 57 2 0 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
51 58 2 0 0 0 0
52 53 1 0 0 0 0
53 54 1 0 0 0 0
54 55 1 0 0 0 0
22 26 1 1 0 0 0
23 27 1 1 0 0 0
31 30 1 0 0 0 0
30 32 1 0 0 0 0
30 33 2 0 0 0 0
27 30 1 0 0 0 0
55 5 1 0 0 0 0
5 4 1 0 0 0 0
5 10 2 0 0 0 0
4 3 1 0 0 0 0
3 2 1 0 0 0 0
3 9 1 6 0 0 0
3 6 1 1 0 0 0
1 8 1 0 0 0 0
1 7 2 0 0 0 0
2 1 1 0 0 0 0
S SKP 7
ID BMFYB5CAa008
NAME (S)-3-Hydroxy-3-methyl-glutaryl-CoA
FORMULA C27H44N7O20P3S
EXACTMASS 911.1574
AVERAGEMASS 911.6607
SMILES C([C@H](C(NCCC(NCCSC(C[C@@](C)(O)CC(O)=O)=O)=O)=O)O)(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)3)O[C@H]([C@@H]3O)n(c21)cnc(c(N)ncn2)1)(O)=O)(O)=O
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C00356
M END