Mol:FLICUNNS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 25 29 0 0 0 0 0 0 0 0999 V2000 | + | 25 29 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.0196 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0196 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0196 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0196 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4633 -0.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4633 -0.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9070 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9070 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9070 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9070 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4633 1.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4633 1.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3507 -0.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3507 -0.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2056 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2056 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2056 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2056 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3507 1.1574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3507 1.1574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7617 -0.1272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7617 -0.1272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7617 -0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7617 -0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3565 -1.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3565 -1.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9512 -0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9512 -0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9512 -0.1272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9512 -0.1272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3565 0.2162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3565 0.2162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6306 1.0347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6306 1.0347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0081 0.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0081 0.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6306 -0.0046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6306 -0.0046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6044 -1.0262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6044 -1.0262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0081 -0.4706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0081 -0.4706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6044 0.0850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6044 0.0850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3507 -0.7694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3507 -0.7694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0472 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0472 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7617 -1.1571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7617 -1.1571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 8 11 1 0 0 0 0 | + | 8 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 11 2 0 0 0 0 | + | 16 11 2 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 1 17 1 0 0 0 0 | + | 1 17 1 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 2 1 0 0 0 0 | + | 19 2 1 0 0 0 0 |
| − | 14 20 1 0 0 0 0 | + | 14 20 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 15 1 0 0 0 0 | + | 22 15 1 0 0 0 0 |
| − | 7 23 1 0 0 0 0 | + | 7 23 1 0 0 0 0 |
| − | 12 24 1 0 0 0 0 | + | 12 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 24 25 | + | M SAL 1 2 24 25 |
| − | M SBL 1 1 28 | + | M SBL 1 1 28 |
| − | M SMT 1 ^OCH3 | + | M SMT 1 ^OCH3 |
| − | M SBV 1 28 -5.9269 2.7619 | + | M SBV 1 28 -5.9269 2.7619 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLICUNNS0001 | + | ID FLICUNNS0001 |
| − | KNApSAcK_ID C00009740 | + | KNApSAcK_ID C00009740 |
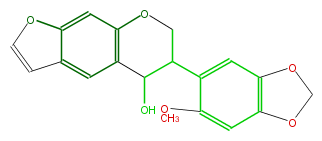
| − | NAME Ambanol | + | NAME Ambanol |
| − | CAS_RN 63838-66-4 | + | CAS_RN 63838-66-4 |
| − | FORMULA C19H16O6 | + | FORMULA C19H16O6 |
| − | EXACTMASS 340.094688244 | + | EXACTMASS 340.094688244 |
| − | AVERAGEMASS 340.32674 | + | AVERAGEMASS 340.32674 |
| − | SMILES c(c45)c(c(OC)cc4OCO5)C(C3O)COc(c32)cc(c1c2)occ1 | + | SMILES c(c45)c(c(OC)cc4OCO5)C(C3O)COc(c32)cc(c1c2)occ1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
25 29 0 0 0 0 0 0 0 0999 V2000
-2.0196 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.0196 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4633 -0.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9070 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9070 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4633 1.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3507 -0.1273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2056 0.1939 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2056 0.8362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3507 1.1574 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7617 -0.1272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7617 -0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3565 -1.1574 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9512 -0.8140 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9512 -0.1272 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3565 0.2162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6306 1.0347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0081 0.5150 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6306 -0.0046 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6044 -1.0262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0081 -0.4706 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6044 0.0850 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3507 -0.7694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0472 -0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7617 -1.1571 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
8 11 1 0 0 0 0
11 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 11 2 0 0 0 0
1 2 1 0 0 0 0
1 17 1 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 2 1 0 0 0 0
14 20 1 0 0 0 0
20 21 1 0 0 0 0
21 22 1 0 0 0 0
22 15 1 0 0 0 0
7 23 1 0 0 0 0
12 24 1 0 0 0 0
24 25 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 24 25
M SBL 1 1 28
M SMT 1 ^OCH3
M SBV 1 28 -5.9269 2.7619
S SKP 8
ID FLICUNNS0001
KNApSAcK_ID C00009740
NAME Ambanol
CAS_RN 63838-66-4
FORMULA C19H16O6
EXACTMASS 340.094688244
AVERAGEMASS 340.32674
SMILES c(c45)c(c(OC)cc4OCO5)C(C3O)COc(c32)cc(c1c2)occ1
M END