Mol:FL7AAAGL0061
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 76 83 0 0 0 0 0 0 0 0999 V2000 | + | 76 83 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.3659 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3659 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3659 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3659 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6514 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6514 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0631 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0631 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0631 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0631 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6514 3.3908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6514 3.3908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7776 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7776 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4920 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4920 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4920 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4920 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7776 3.3908 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 | + | 0.7776 3.3908 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2686 3.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2686 3.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9830 3.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9830 3.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6975 3.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6975 3.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6975 4.2516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6975 4.2516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9830 4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9830 4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2686 4.2516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2686 4.2516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3377 4.6212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3377 4.6212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9458 3.3131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9458 3.3131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6514 0.9781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6514 0.9781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1422 1.7779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1422 1.7779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1139 1.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1139 1.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7013 0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7013 0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9029 0.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9029 0.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1095 0.2779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1095 0.2779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5220 0.9927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.5220 0.9927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3204 0.7836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3204 0.7836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4774 1.3694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4774 1.3694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9573 1.6465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9573 1.6465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5971 0.5271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5971 0.5271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1500 0.5546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1500 0.5546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0744 -0.1352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0744 -0.1352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9660 -0.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9660 -0.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7911 -0.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7911 -0.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0256 0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0256 0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6305 1.0184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6305 1.0184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8054 1.0355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8054 1.0355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5709 0.2442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5709 0.2442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9884 0.4133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9884 0.4133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5029 0.1462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5029 0.1462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1292 -0.9808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1292 -0.9808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3372 -0.5840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3372 -0.5840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4842 -0.0210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4842 -0.0210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1959 -0.7611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1959 -0.7611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6216 -1.4984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6216 -1.4984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6280 -0.7611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6280 -0.7611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0399 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0399 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8638 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8638 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2763 -0.7601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2763 -0.7601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1013 -0.7601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1013 -0.7601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5138 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5138 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1013 -2.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1013 -2.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.2763 -2.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2763 -2.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3377 -1.4746 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3377 -1.4746 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7859 -2.4667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7859 -2.4667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5901 -2.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5901 -2.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6240 -1.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6240 -1.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0733 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0733 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2690 -0.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2690 -0.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2351 -1.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2351 -1.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6290 -1.7530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6290 -1.7530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2235 -2.1306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2235 -2.1306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1064 -3.0703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1064 -3.0703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2110 -2.6347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2110 -2.6347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1856 -1.8084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1856 -1.8084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7052 -3.2362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7052 -3.2362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0982 -3.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0982 -3.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1187 -3.2362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1187 -3.2362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5306 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5306 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3545 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3545 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7670 -3.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7670 -3.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5920 -3.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5920 -3.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0045 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0045 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5920 -4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5920 -4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7670 -4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7670 -4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8284 -3.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8284 -3.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0040 -2.5217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0040 -2.5217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 14 17 1 0 0 0 0 | + | 14 17 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 21 29 1 0 0 0 0 | + | 21 29 1 0 0 0 0 |
| − | 22 30 1 0 0 0 0 | + | 22 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 32 40 1 0 0 0 0 | + | 32 40 1 0 0 0 0 |
| − | 33 41 1 0 0 0 0 | + | 33 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 20 1 0 0 0 0 | + | 35 20 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 47 1 0 0 0 0 | + | 52 47 1 0 0 0 0 |
| − | 50 53 1 0 0 0 0 | + | 50 53 1 0 0 0 0 |
| − | 54 55 1 1 0 0 0 | + | 54 55 1 1 0 0 0 |
| − | 55 56 1 1 0 0 0 | + | 55 56 1 1 0 0 0 |
| − | 57 56 1 1 0 0 0 | + | 57 56 1 1 0 0 0 |
| − | 57 58 1 0 0 0 0 | + | 57 58 1 0 0 0 0 |
| − | 58 59 1 0 0 0 0 | + | 58 59 1 0 0 0 0 |
| − | 59 54 1 0 0 0 0 | + | 59 54 1 0 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 60 61 1 0 0 0 0 | + | 60 61 1 0 0 0 0 |
| − | 54 62 1 0 0 0 0 | + | 54 62 1 0 0 0 0 |
| − | 55 63 1 0 0 0 0 | + | 55 63 1 0 0 0 0 |
| − | 56 64 1 0 0 0 0 | + | 56 64 1 0 0 0 0 |
| − | 57 42 1 0 0 0 0 | + | 57 42 1 0 0 0 0 |
| − | 43 39 1 0 0 0 0 | + | 43 39 1 0 0 0 0 |
| − | 65 66 2 0 0 0 0 | + | 65 66 2 0 0 0 0 |
| − | 65 67 1 0 0 0 0 | + | 65 67 1 0 0 0 0 |
| − | 67 68 2 0 0 0 0 | + | 67 68 2 0 0 0 0 |
| − | 68 69 1 0 0 0 0 | + | 68 69 1 0 0 0 0 |
| − | 69 70 2 0 0 0 0 | + | 69 70 2 0 0 0 0 |
| − | 70 71 1 0 0 0 0 | + | 70 71 1 0 0 0 0 |
| − | 71 72 2 0 0 0 0 | + | 71 72 2 0 0 0 0 |
| − | 72 73 1 0 0 0 0 | + | 72 73 1 0 0 0 0 |
| − | 73 74 2 0 0 0 0 | + | 73 74 2 0 0 0 0 |
| − | 74 69 1 0 0 0 0 | + | 74 69 1 0 0 0 0 |
| − | 72 75 1 0 0 0 0 | + | 72 75 1 0 0 0 0 |
| − | 71 76 1 0 0 0 0 | + | 71 76 1 0 0 0 0 |
| − | 61 65 1 0 0 0 0 | + | 61 65 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL7AAAGL0061 | + | ID FL7AAAGL0061 |
| − | KNApSAcK_ID C00014837 | + | KNApSAcK_ID C00014837 |
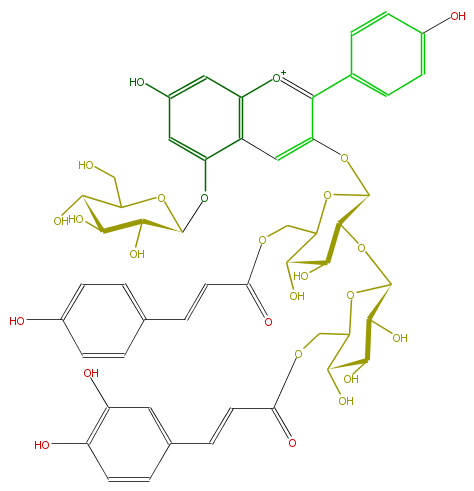
| − | NAME Pelargonidin 3-O-[2-O-(6-(E)-caffeoyl-beta-D-glucopyranosyl)-6-O-(E)-p-coumaroyl-beta-D-glucopyranoside]-5-O-(beta-D-glucopyranoside);Pelargonidin 3-(2-(6-caffeylglucosyl)-6-p-coumarylglucoside)-5-glucoside | + | NAME Pelargonidin 3-O-[2-O-(6-(E)-caffeoyl-beta-D-glucopyranosyl)-6-O-(E)-p-coumaroyl-beta-D-glucopyranoside]-5-O-(beta-D-glucopyranoside);Pelargonidin 3-(2-(6-caffeylglucosyl)-6-p-coumarylglucoside)-5-glucoside |
| − | CAS_RN 448963-06-2 | + | CAS_RN 448963-06-2 |
| − | FORMULA C51H53O25 | + | FORMULA C51H53O25 |
| − | EXACTMASS 1065.287592246 | + | EXACTMASS 1065.287592246 |
| − | AVERAGEMASS 1065.95152 | + | AVERAGEMASS 1065.95152 |
| − | SMILES C(C8O)(O)C(OC(C8O)COC(C=Cc(c7)ccc(O)c(O)7)=O)OC(C6O)C(OC(C6O)COC(=O)C=Cc(c5)ccc(O)c5)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)1)c(OC(O2)C(O)C(C(O)C2CO)O)cc(O)c1 | + | SMILES C(C8O)(O)C(OC(C8O)COC(C=Cc(c7)ccc(O)c(O)7)=O)OC(C6O)C(OC(C6O)COC(=O)C=Cc(c5)ccc(O)c5)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)1)c(OC(O2)C(O)C(C(O)C2CO)O)cc(O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
76 83 0 0 0 0 0 0 0 0999 V2000
-1.3659 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3659 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6514 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0631 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0631 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6514 3.3908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7776 1.7408 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4920 2.1533 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4920 2.9783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7776 3.3908 0.0000 O 0 3 0 0 0 0 0 0 0 0 0 0
2.2686 3.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9830 3.0141 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6975 3.4266 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6975 4.2516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9830 4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2686 4.2516 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3377 4.6212 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9458 3.3131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6514 0.9781 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1422 1.7779 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1139 1.0103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7013 0.2956 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9029 0.5048 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1095 0.2779 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.5220 0.9927 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3204 0.7836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4774 1.3694 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9573 1.6465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.5971 0.5271 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1500 0.5546 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0744 -0.1352 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9660 -0.3172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7911 -0.3343 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0256 0.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6305 1.0184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8054 1.0355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5709 0.2442 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9884 0.4133 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5029 0.1462 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1292 -0.9808 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3372 -0.5840 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4842 -0.0210 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1959 -0.7611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6216 -1.4984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6280 -0.7611 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0399 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8638 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2763 -0.7601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1013 -0.7601 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5138 -1.4746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1013 -2.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.2763 -2.1891 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3377 -1.4746 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7859 -2.4667 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5901 -2.2816 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6240 -1.4570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0733 -0.7647 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2690 -0.9498 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2351 -1.7745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6290 -1.7530 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2235 -2.1306 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1064 -3.0703 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2110 -2.6347 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1856 -1.8084 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7052 -3.2362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0982 -3.9169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1187 -3.2362 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5306 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3545 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7670 -3.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5920 -3.2352 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0045 -3.9497 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5920 -4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7670 -4.6641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8284 -3.9497 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0040 -2.5217 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 2 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 5 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
14 17 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
8 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
21 29 1 0 0 0 0
22 30 1 0 0 0 0
23 31 1 0 0 0 0
24 19 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
32 40 1 0 0 0 0
33 41 1 0 0 0 0
34 42 1 0 0 0 0
35 20 1 0 0 0 0
43 44 2 0 0 0 0
43 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 47 1 0 0 0 0
50 53 1 0 0 0 0
54 55 1 1 0 0 0
55 56 1 1 0 0 0
57 56 1 1 0 0 0
57 58 1 0 0 0 0
58 59 1 0 0 0 0
59 54 1 0 0 0 0
59 60 1 0 0 0 0
60 61 1 0 0 0 0
54 62 1 0 0 0 0
55 63 1 0 0 0 0
56 64 1 0 0 0 0
57 42 1 0 0 0 0
43 39 1 0 0 0 0
65 66 2 0 0 0 0
65 67 1 0 0 0 0
67 68 2 0 0 0 0
68 69 1 0 0 0 0
69 70 2 0 0 0 0
70 71 1 0 0 0 0
71 72 2 0 0 0 0
72 73 1 0 0 0 0
73 74 2 0 0 0 0
74 69 1 0 0 0 0
72 75 1 0 0 0 0
71 76 1 0 0 0 0
61 65 1 0 0 0 0
S SKP 8
ID FL7AAAGL0061
KNApSAcK_ID C00014837
NAME Pelargonidin 3-O-[2-O-(6-(E)-caffeoyl-beta-D-glucopyranosyl)-6-O-(E)-p-coumaroyl-beta-D-glucopyranoside]-5-O-(beta-D-glucopyranoside);Pelargonidin 3-(2-(6-caffeylglucosyl)-6-p-coumarylglucoside)-5-glucoside
CAS_RN 448963-06-2
FORMULA C51H53O25
EXACTMASS 1065.287592246
AVERAGEMASS 1065.95152
SMILES C(C8O)(O)C(OC(C8O)COC(C=Cc(c7)ccc(O)c(O)7)=O)OC(C6O)C(OC(C6O)COC(=O)C=Cc(c5)ccc(O)c5)Oc(c3c(c4)ccc(c4)O)cc(c([o+1]3)1)c(OC(O2)C(O)C(C(O)C2CO)O)cc(O)c1
M END