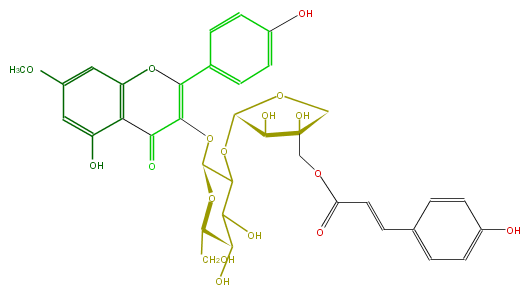
Mol:FL5FCAGL0008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 53 58 0 0 0 0 0 0 0 0999 V2000 | + | 53 58 0 0 0 0 0 0 0 0999 V2000 |
| − | -4.7626 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7626 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7626 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7626 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0515 0.6398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0515 0.6398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3405 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3405 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3405 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3405 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0515 2.2818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0515 2.2818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6294 0.6398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6294 0.6398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9184 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9184 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9184 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9184 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6294 2.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6294 2.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6294 -0.0936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6294 -0.0936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2087 2.3161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2087 2.3161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4840 1.8977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4840 1.8977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2406 2.3161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2406 2.3161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2406 3.1529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2406 3.1529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4840 3.5713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4840 3.5713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2087 3.1529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2087 3.1529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0515 -0.0730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0515 -0.0730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0164 3.6007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0164 3.6007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2164 0.5670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2164 0.5670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6778 -0.4947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6778 -0.4947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3850 -0.0865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3850 -0.0865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1606 -0.8716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1606 -0.8716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3850 -1.6615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3850 -1.6615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6778 -2.0699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6778 -2.0699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9021 -1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9021 -1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8811 0.2635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8811 0.2635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2153 -1.7755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2153 -1.7755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9897 -2.9039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9897 -2.9039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6073 1.1353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6073 1.1353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1288 0.6321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1288 0.6321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9492 0.6908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9492 0.6908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6711 1.2064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6711 1.2064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4955 1.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4955 1.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1288 1.1260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1288 1.1260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9492 0.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9492 0.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9492 1.1445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9492 1.1445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4132 -0.2788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4132 -0.2788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8774 -0.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8774 -0.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4668 -1.6959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4668 -1.6959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6120 -0.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6120 -0.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0015 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0015 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7247 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7247 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1373 -2.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1373 -2.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9624 -2.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9624 -2.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.3749 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.3749 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9624 -0.9445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9624 -0.9445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1373 -0.9445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1373 -0.9445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.0645 -1.6590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.0645 -1.6590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.4255 2.2540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.4255 2.2540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.0645 3.3610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.0645 3.3610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4369 -2.3642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4369 -2.3642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7608 -3.6007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7608 -3.6007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 20 8 1 0 0 0 0 | + | 20 8 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 22 20 1 0 0 0 0 | + | 22 20 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 30 27 1 0 0 0 0 | + | 30 27 1 0 0 0 0 |
| − | 36 38 1 0 0 0 0 | + | 36 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 39 41 1 0 0 0 0 | + | 39 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 43 44 2 0 0 0 0 | + | 43 44 2 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 46 49 1 0 0 0 0 | + | 46 49 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 1 50 1 0 0 0 0 | + | 1 50 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 24 52 1 0 0 0 0 | + | 24 52 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 50 51 | + | M SAL 1 2 50 51 |
| − | M SBL 1 1 56 | + | M SBL 1 1 56 |
| − | M SMT 1 ^ OCH3 | + | M SMT 1 ^ OCH3 |
| − | M SBV 1 56 0.6630 -0.3828 | + | M SBV 1 56 0.6630 -0.3828 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 52 53 | + | M SAL 2 2 52 53 |
| − | M SBL 2 1 58 | + | M SBL 2 1 58 |
| − | M SMT 2 ^ CH2OH | + | M SMT 2 ^ CH2OH |
| − | M SBV 2 58 0.0518 0.7027 | + | M SBV 2 58 0.0518 0.7027 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FCAGL0008 | + | ID FL5FCAGL0008 |
| − | FORMULA C36H36O17 | + | FORMULA C36H36O17 |
| − | EXACTMASS 740.1952497259999 | + | EXACTMASS 740.1952497259999 |
| − | AVERAGEMASS 740.66084 | + | AVERAGEMASS 740.66084 |
| − | SMILES c(c12)(cc(cc1OC(c(c6)ccc(c6)O)=C(OC(C3OC(C(O)4)OCC(O)(COC(C=Cc(c5)ccc(O)c5)=O)4)OC(CO)C(C(O)3)O)C2=O)OC)O | + | SMILES c(c12)(cc(cc1OC(c(c6)ccc(c6)O)=C(OC(C3OC(C(O)4)OCC(O)(COC(C=Cc(c5)ccc(O)c5)=O)4)OC(CO)C(C(O)3)O)C2=O)OC)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
53 58 0 0 0 0 0 0 0 0999 V2000
-4.7626 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.7626 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0515 0.6398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3405 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3405 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0515 2.2818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6294 0.6398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9184 1.0503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9184 1.8713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6294 2.2818 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6294 -0.0936 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2087 2.3161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4840 1.8977 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2406 2.3161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2406 3.1529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4840 3.5713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2087 3.1529 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0515 -0.0730 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0164 3.6007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2164 0.5670 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6778 -0.4947 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3850 -0.0865 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1606 -0.8716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3850 -1.6615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6778 -2.0699 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.9021 -1.2846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8811 0.2635 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2153 -1.7755 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9897 -2.9039 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6073 1.1353 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1288 0.6321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9492 0.6908 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6711 1.2064 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4955 1.6189 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1288 1.1260 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9492 0.1545 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9492 1.1445 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4132 -0.2788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8774 -0.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4668 -1.6959 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6120 -0.9845 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0015 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7247 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1373 -2.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9624 -2.3736 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.3749 -1.6590 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9624 -0.9445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1373 -0.9445 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.0645 -1.6590 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.4255 2.2540 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-6.0645 3.3610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4369 -2.3642 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7608 -3.6007 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
20 8 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
22 20 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 30 1 0 0 0 0
31 35 1 0 0 0 0
32 36 1 0 0 0 0
32 37 1 0 0 0 0
30 27 1 0 0 0 0
36 38 1 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
39 41 1 0 0 0 0
41 42 2 0 0 0 0
42 43 1 0 0 0 0
43 44 2 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
46 47 1 0 0 0 0
47 48 2 0 0 0 0
48 43 1 0 0 0 0
46 49 1 0 0 0 0
50 51 1 0 0 0 0
1 50 1 0 0 0 0
52 53 1 0 0 0 0
24 52 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 50 51
M SBL 1 1 56
M SMT 1 ^ OCH3
M SBV 1 56 0.6630 -0.3828
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 52 53
M SBL 2 1 58
M SMT 2 ^ CH2OH
M SBV 2 58 0.0518 0.7027
S SKP 5
ID FL5FCAGL0008
FORMULA C36H36O17
EXACTMASS 740.1952497259999
AVERAGEMASS 740.66084
SMILES c(c12)(cc(cc1OC(c(c6)ccc(c6)O)=C(OC(C3OC(C(O)4)OCC(O)(COC(C=Cc(c5)ccc(O)c5)=O)4)OC(CO)C(C(O)3)O)C2=O)OC)O
M END