Mol:FL5FALGS0002
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 40 0 0 0 0 0 0 0 0999 V2000 | + | 37 40 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3956 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3956 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3956 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3956 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8393 -1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8393 -1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2830 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2830 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2830 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2830 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8393 -0.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8393 -0.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7267 -1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7267 -1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1704 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1704 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1704 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1704 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7267 -0.1551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7267 -0.1551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7267 -1.9406 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7267 -1.9406 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3857 -0.1552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3857 -0.1552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9527 -0.4825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9527 -0.4825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5197 -0.1552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5197 -0.1552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5197 0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5197 0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9527 0.8268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9527 0.8268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3857 0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3857 0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8393 -2.0819 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8393 -2.0819 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0865 -0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0865 -0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8183 1.7801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8183 1.7801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0081 1.8760 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -2.0081 1.8760 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.9202 1.1745 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.9202 1.1745 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.2682 1.0741 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.2682 1.0741 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7512 0.7831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.7512 0.7831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.9090 1.3720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9090 1.3720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.5656 1.4907 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.5656 1.4907 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.3685 1.4333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3685 1.4333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2588 0.3859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2588 0.3859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1214 1.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1214 1.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2561 2.0819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2561 2.0819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5416 1.6694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5416 1.6694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6956 -1.6186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6956 -1.6186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5616 -2.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5616 -2.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1038 0.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1038 0.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8183 0.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8183 0.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7528 0.1425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7528 0.1425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2528 1.0085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2528 1.0085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 14 19 1 0 0 0 0 | + | 14 19 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 22 27 1 0 0 0 0 | + | 22 27 1 0 0 0 0 |
| − | 23 28 1 0 0 0 0 | + | 23 28 1 0 0 0 0 |
| − | 20 21 1 0 0 0 0 | + | 20 21 1 0 0 0 0 |
| − | 17 29 1 0 0 0 0 | + | 17 29 1 0 0 0 0 |
| − | 29 24 1 0 0 0 0 | + | 29 24 1 0 0 0 0 |
| − | 26 30 1 0 0 0 0 | + | 26 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 8 32 1 0 0 0 0 | + | 8 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 15 34 1 0 0 0 0 | + | 15 34 1 0 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 1 36 1 0 0 0 0 | + | 1 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | M STY 1 4 SUP | + | M STY 1 4 SUP |
| − | M SLB 1 4 4 | + | M SLB 1 4 4 |
| − | M SAL 4 2 30 31 | + | M SAL 4 2 30 31 |
| − | M SBL 4 1 33 | + | M SBL 4 1 33 |
| − | M SMT 4 CH2OH | + | M SMT 4 CH2OH |
| − | M SVB 4 33 -1.2561 2.0819 | + | M SVB 4 33 -1.2561 2.0819 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 36 37 | + | M SAL 3 2 36 37 |
| − | M SBL 3 1 39 | + | M SBL 3 1 39 |
| − | M SMT 3 OCH3 | + | M SMT 3 OCH3 |
| − | M SVB 3 39 -2.7528 0.1425 | + | M SVB 3 39 -2.7528 0.1425 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 34 35 | + | M SAL 2 2 34 35 |
| − | M SBL 2 1 37 | + | M SBL 2 1 37 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SVB 2 37 2.1038 0.8184 | + | M SVB 2 37 2.1038 0.8184 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 32 33 | + | M SAL 1 2 32 33 |
| − | M SBL 1 1 35 | + | M SBL 1 1 35 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 35 0.0285 -1.3249 | + | M SVB 1 35 0.0285 -1.3249 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FALGS0002 | + | ID FL5FALGS0002 |
| − | KNApSAcK_ID C00005723 | + | KNApSAcK_ID C00005723 |
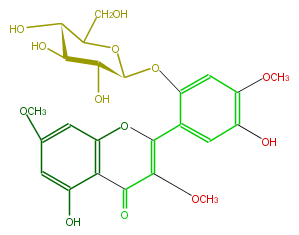
| − | NAME Chrysosplenoside A | + | NAME Chrysosplenoside A |
| − | CAS_RN 23615-30-7 | + | CAS_RN 23615-30-7 |
| − | FORMULA C24H26O13 | + | FORMULA C24H26O13 |
| − | EXACTMASS 522.137340918 | + | EXACTMASS 522.137340918 |
| − | AVERAGEMASS 522.45544 | + | AVERAGEMASS 522.45544 |
| − | SMILES c(c1OC)(O)cc(C(=C4OC)Oc(c3)c(C(=O)4)c(O)cc(OC)3)c(O[C@@H]([C@H]2O)OC([C@H](O)[C@@H]2O)CO)c1 | + | SMILES c(c1OC)(O)cc(C(=C4OC)Oc(c3)c(C(=O)4)c(O)cc(OC)3)c(O[C@@H]([C@H]2O)OC([C@H](O)[C@@H]2O)CO)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 40 0 0 0 0 0 0 0 0999 V2000
-2.3956 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3956 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8393 -1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2830 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2830 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8393 -0.1551 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7267 -1.4398 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1704 -1.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1704 -0.4762 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7267 -0.1551 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7267 -1.9406 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3857 -0.1552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9527 -0.4825 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5197 -0.1552 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5197 0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9527 0.8268 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3857 0.4995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8393 -2.0819 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0865 -0.4824 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8183 1.7801 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0081 1.8760 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.9202 1.1745 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.2682 1.0741 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7512 0.7831 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9090 1.3720 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.5656 1.4907 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.3685 1.4333 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2588 0.3859 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1214 1.0067 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.2561 2.0819 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5416 1.6694 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6956 -1.6186 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5616 -2.1186 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1038 0.8184 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8183 0.4059 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7528 0.1425 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.2528 1.0085 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
14 19 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
22 27 1 0 0 0 0
23 28 1 0 0 0 0
20 21 1 0 0 0 0
17 29 1 0 0 0 0
29 24 1 0 0 0 0
26 30 1 0 0 0 0
30 31 1 0 0 0 0
8 32 1 0 0 0 0
32 33 1 0 0 0 0
15 34 1 0 0 0 0
34 35 1 0 0 0 0
1 36 1 0 0 0 0
36 37 1 0 0 0 0
M STY 1 4 SUP
M SLB 1 4 4
M SAL 4 2 30 31
M SBL 4 1 33
M SMT 4 CH2OH
M SVB 4 33 -1.2561 2.0819
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 36 37
M SBL 3 1 39
M SMT 3 OCH3
M SVB 3 39 -2.7528 0.1425
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 34 35
M SBL 2 1 37
M SMT 2 OCH3
M SVB 2 37 2.1038 0.8184
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 32 33
M SBL 1 1 35
M SMT 1 OCH3
M SVB 1 35 0.0285 -1.3249
S SKP 8
ID FL5FALGS0002
KNApSAcK_ID C00005723
NAME Chrysosplenoside A
CAS_RN 23615-30-7
FORMULA C24H26O13
EXACTMASS 522.137340918
AVERAGEMASS 522.45544
SMILES c(c1OC)(O)cc(C(=C4OC)Oc(c3)c(C(=O)4)c(O)cc(OC)3)c(O[C@@H]([C@H]2O)OC([C@H](O)[C@@H]2O)CO)c1
M END