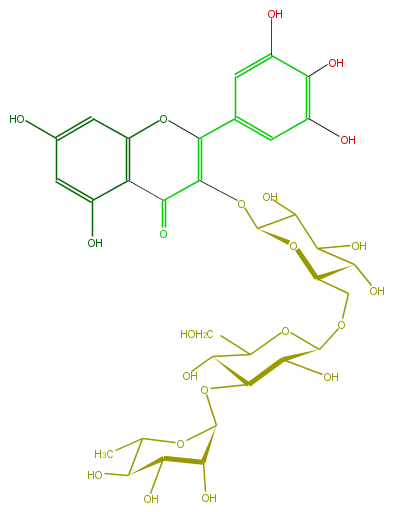
Mol:FL5FAGGL0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.8189 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8189 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8189 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8189 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1040 1.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1040 1.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3890 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3890 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3890 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3890 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1040 2.7506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1040 2.7506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 1.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 1.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0408 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0408 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0408 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0408 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6741 2.7506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6741 2.7506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6742 0.4559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6742 0.4559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7555 2.7504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7555 2.7504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4841 2.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4841 2.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2128 2.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2128 2.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2128 3.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2128 3.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4842 4.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4842 4.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7555 3.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7555 3.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5336 2.7504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5336 2.7504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1038 0.2745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1038 0.2745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7007 3.8734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7007 3.8734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9490 0.8198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9490 0.8198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1925 0.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1925 0.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9278 0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9278 0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3820 -0.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3820 -0.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1387 -0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1387 -0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4035 0.1498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4035 0.1498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3962 1.1835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3962 1.1835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1627 0.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1627 0.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5336 -0.6918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5336 -0.6918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0138 -0.7494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0138 -0.7494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3026 -2.0211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3026 -2.0211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0199 -2.5787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0199 -2.5787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7036 -2.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7036 -2.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4377 -1.8723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4377 -1.8723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7593 -1.4804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7593 -1.4804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0548 -1.9692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0548 -1.9692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2203 -2.3778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2203 -2.3778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1452 -2.6665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1452 -2.6665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6084 -2.4085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6084 -2.4085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8798 -1.3640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8798 -1.3640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8856 1.0543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8856 1.0543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4841 4.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4841 4.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1135 -3.6588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1135 -3.6588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3814 -4.3953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3814 -4.3953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6769 -4.0524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6769 -4.0524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1068 -4.1328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1068 -4.1328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3747 -3.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3747 -3.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3298 -3.7394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3298 -3.7394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7535 -3.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7535 -3.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9774 -4.3575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9774 -4.3575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9844 -4.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9844 -4.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1610 -4.8263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1610 -4.8263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9412 2.3298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9412 2.3298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3697 -1.5480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3697 -1.5480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5112 -0.6121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5112 -0.6121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 20 15 1 0 0 0 0 | + | 20 15 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 26 28 1 0 0 0 0 | + | 26 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 40 30 1 0 0 0 0 | + | 40 30 1 0 0 0 0 |
| − | 22 41 1 0 0 0 0 | + | 22 41 1 0 0 0 0 |
| − | 41 8 1 0 0 0 0 | + | 41 8 1 0 0 0 0 |
| − | 16 42 1 0 0 0 0 | + | 16 42 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 47 46 1 1 0 0 0 | + | 47 46 1 1 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 47 38 1 0 0 0 0 | + | 47 38 1 0 0 0 0 |
| − | 14 53 1 0 0 0 0 | + | 14 53 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 36 54 1 0 0 0 0 | + | 36 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 54 55 | + | M SAL 1 2 54 55 |
| − | M SBL 1 1 60 | + | M SBL 1 1 60 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 60 0.6851 -0.4212 | + | M SBV 1 60 0.6851 -0.4212 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAGGL0007 | + | ID FL5FAGGL0007 |
| − | FORMULA C33H40O22 | + | FORMULA C33H40O22 |
| − | EXACTMASS 788.201122964 | + | EXACTMASS 788.201122964 |
| − | AVERAGEMASS 788.6575 | + | AVERAGEMASS 788.6575 |
| − | SMILES C(C(O)1)(C(O)C(OC1OC(C6O)C(C(OC6CO)OCC(O2)C(O)C(O)C(C(OC(C(=O)3)=C(c(c5)cc(c(O)c(O)5)O)Oc(c4)c3c(O)cc4O)2)O)O)C)O | + | SMILES C(C(O)1)(C(O)C(OC1OC(C6O)C(C(OC6CO)OCC(O2)C(O)C(O)C(C(OC(C(=O)3)=C(c(c5)cc(c(O)c(O)5)O)Oc(c4)c3c(O)cc4O)2)O)O)C)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-2.8189 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8189 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1040 1.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3890 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3890 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1040 2.7506 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 1.0995 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0408 1.5122 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0408 2.3378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6741 2.7506 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6742 0.4559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7555 2.7504 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4841 2.3298 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2128 2.7503 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2128 3.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4842 4.0124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7555 3.5918 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5336 2.7504 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1038 0.2745 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7007 3.8734 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9490 0.8198 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1925 0.5444 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9278 0.2164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3820 -0.4537 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1387 -0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4035 0.1498 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3962 1.1835 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1627 0.2045 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5336 -0.6918 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0138 -0.7494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3026 -2.0211 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0199 -2.5787 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7036 -2.0775 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4377 -1.8723 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7593 -1.4804 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0548 -1.9692 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2203 -2.3778 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1452 -2.6665 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6084 -2.4085 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8798 -1.3640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.8856 1.0543 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4841 4.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1135 -3.6588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3814 -4.3953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6769 -4.0524 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1068 -4.1328 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3747 -3.3966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3298 -3.7394 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7535 -3.9373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9774 -4.3575 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9844 -4.8534 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1610 -4.8263 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9412 2.3298 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3697 -1.5480 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5112 -0.6121 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
3 19 1 0 0 0 0
20 15 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
26 28 1 0 0 0 0
25 29 1 0 0 0 0
24 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
40 30 1 0 0 0 0
22 41 1 0 0 0 0
41 8 1 0 0 0 0
16 42 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 1 0 0 0
45 46 1 1 0 0 0
47 46 1 1 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 52 1 0 0 0 0
47 38 1 0 0 0 0
14 53 1 0 0 0 0
54 55 1 0 0 0 0
36 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 54 55
M SBL 1 1 60
M SMT 1 ^ CH2OH
M SBV 1 60 0.6851 -0.4212
S SKP 5
ID FL5FAGGL0007
FORMULA C33H40O22
EXACTMASS 788.201122964
AVERAGEMASS 788.6575
SMILES C(C(O)1)(C(O)C(OC1OC(C6O)C(C(OC6CO)OCC(O2)C(O)C(O)C(C(OC(C(=O)3)=C(c(c5)cc(c(O)c(O)5)O)Oc(c4)c3c(O)cc4O)2)O)O)C)O
M END