Mol:FL5FADGL0040
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | 0.0739 0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0739 0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7007 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7007 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0739 2.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0739 2.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5528 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5528 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5528 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5528 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3275 0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3275 0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9543 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9543 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9543 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9543 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3275 2.3761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3275 2.3761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5837 2.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5837 2.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1947 2.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1947 2.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8058 2.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8058 2.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8058 3.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8058 3.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1947 3.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1947 3.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5837 3.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5837 3.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3275 0.3344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3275 0.3344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2479 3.3384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2479 3.3384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1947 3.9490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1947 3.9490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0739 0.3996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0739 0.3996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4346 1.0131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4346 1.0131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0741 2.3151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0741 2.3151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7978 2.4106 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.7978 2.4106 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.2184 1.8234 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.2184 1.8234 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.4989 2.2271 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.4989 2.2271 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -1.6741 2.2078 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.6741 2.2078 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.2535 2.7951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.2535 2.7951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9730 2.3915 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -2.9730 2.3915 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -3.2561 2.8817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2561 2.8817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.9378 3.2000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.9378 3.2000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2479 1.9605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2479 1.9605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7131 1.9559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7131 1.9559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3717 1.7525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3717 1.7525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1669 -1.0655 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 1.1669 -1.0655 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 1.9887 -1.1386 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 1.9887 -1.1386 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.2763 -0.3653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2763 -0.3653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.9178 0.1535 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.9178 0.1535 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.0960 0.2267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0960 0.2267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8083 -0.5466 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 1.8083 -0.5466 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.7796 -0.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7796 -0.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1669 -1.7117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1669 -1.7117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5426 -1.3961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5426 -1.3961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2611 -0.2307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2611 -0.2307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7318 -0.5362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7318 -0.5362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1993 -1.0688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1993 -1.0688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3532 -0.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3532 -0.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1993 -1.6620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1993 -1.6620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -2.0206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -2.0206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -2.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -2.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0030 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0030 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0030 -3.5927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0030 -3.5927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -3.9282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -3.9282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1592 -3.5927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1592 -3.5927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1592 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1592 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4219 -4.4177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4219 -4.4177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6635 4.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6635 4.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 3 1 0 0 0 0 | + | 10 3 1 0 0 0 0 |
| − | 9 11 1 0 0 0 0 | + | 9 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 5 22 1 0 0 0 0 | + | 5 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 22 26 1 0 0 0 0 | + | 22 26 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 21 37 1 0 0 0 0 | + | 21 37 1 0 0 0 0 |
| − | 36 40 1 0 0 0 0 | + | 36 40 1 0 0 0 0 |
| − | 34 41 1 0 0 0 0 | + | 34 41 1 0 0 0 0 |
| − | 35 42 1 0 0 0 0 | + | 35 42 1 0 0 0 0 |
| − | 39 43 1 0 0 0 0 | + | 39 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 44 45 1 0 0 0 0 | + | 44 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 47 48 2 0 0 0 0 | + | 47 48 2 0 0 0 0 |
| − | 19 56 1 0 0 0 0 | + | 19 56 1 0 0 0 0 |
| − | 52 55 1 0 0 0 0 | + | 52 55 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 2 0 0 0 0 | + | 49 50 2 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 2 0 0 0 0 | + | 51 52 2 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 53 54 2 0 0 0 0 | + | 53 54 2 0 0 0 0 |
| − | 54 49 1 0 0 0 0 | + | 54 49 1 0 0 0 0 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 29 30 | + | M SAL 2 2 29 30 |
| − | M SBL 2 1 32 | + | M SBL 2 1 32 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 32 -3.2561 2.8817 | + | M SVB 2 32 -3.2561 2.8817 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 19 56 | + | M SAL 1 2 19 56 |
| − | M SBL 1 1 21 | + | M SBL 1 1 21 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 21 3.1947 3.949 | + | M SVB 1 21 3.1947 3.949 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FADGL0040 | + | ID FL5FADGL0040 |
| − | KNApSAcK_ID C00011130 | + | KNApSAcK_ID C00011130 |
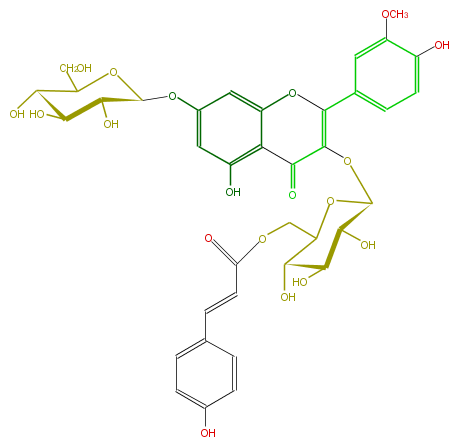
| − | NAME Isorhamnetin 3-O-beta-(6''-O-E-p-coumaroylglucoside)-7-O-beta-glucoside;7-(beta-D-Glucopyranosyloxy)-5-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-3-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one | + | NAME Isorhamnetin 3-O-beta-(6''-O-E-p-coumaroylglucoside)-7-O-beta-glucoside;7-(beta-D-Glucopyranosyloxy)-5-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-3-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one |
| − | CAS_RN 473919-26-5 | + | CAS_RN 473919-26-5 |
| − | FORMULA C37H38O19 | + | FORMULA C37H38O19 |
| − | EXACTMASS 786.200729034 | + | EXACTMASS 786.200729034 |
| − | AVERAGEMASS 786.68622 | + | AVERAGEMASS 786.68622 |
| − | SMILES O[C@@H]([C@@H](O)6)[C@@H](OC(CO)[C@H](O)6)Oc(c5)cc(c1c5O)OC(c(c4)ccc(O)c4OC)=C(O[C@H](O2)[C@@H](O)[C@H]([C@H](C(COC(=O)C=Cc(c3)ccc(c3)O)2)O)O)C(=O)1 | + | SMILES O[C@@H]([C@@H](O)6)[C@@H](OC(CO)[C@H](O)6)Oc(c5)cc(c1c5O)OC(c(c4)ccc(O)c4OC)=C(O[C@H](O2)[C@@H](O)[C@H]([C@H](C(COC(=O)C=Cc(c3)ccc(c3)O)2)O)O)C(=O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
0.0739 0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7007 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0739 2.3761 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5528 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5528 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3275 0.9286 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9543 1.2905 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9543 2.0142 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3275 2.3761 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5837 2.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1947 2.0248 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8058 2.3776 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8058 3.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1947 3.4360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5837 3.0832 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3275 0.3344 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2479 3.3384 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1947 3.9490 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0739 0.3996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4346 1.0131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.0741 2.3151 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7978 2.4106 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.2184 1.8234 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.4989 2.2271 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-1.6741 2.2078 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.2535 2.7951 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.9730 2.3915 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-3.2561 2.8817 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.9378 3.2000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2479 1.9605 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7131 1.9559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3717 1.7525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1669 -1.0655 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
1.9887 -1.1386 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.2763 -0.3653 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.9178 0.1535 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.0960 0.2267 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8083 -0.5466 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.7796 -0.6559 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1669 -1.7117 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5426 -1.3961 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2611 -0.2307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7318 -0.5362 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1993 -1.0688 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3532 -0.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.1993 -1.6620 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -2.0206 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -2.5862 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0030 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0030 -3.5927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -3.9282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1592 -3.5927 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1592 -2.9217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4219 -4.4177 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6635 4.4177 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 3 1 0 0 0 0
9 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
14 18 1 0 0 0 0
15 19 1 0 0 0 0
1 20 1 0 0 0 0
8 21 1 0 0 0 0
5 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
22 26 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
21 37 1 0 0 0 0
36 40 1 0 0 0 0
34 41 1 0 0 0 0
35 42 1 0 0 0 0
39 43 1 0 0 0 0
43 44 1 0 0 0 0
44 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
47 48 2 0 0 0 0
19 56 1 0 0 0 0
52 55 1 0 0 0 0
48 49 1 0 0 0 0
49 50 2 0 0 0 0
50 51 1 0 0 0 0
51 52 2 0 0 0 0
52 53 1 0 0 0 0
53 54 2 0 0 0 0
54 49 1 0 0 0 0
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 29 30
M SBL 2 1 32
M SMT 2 CH2OH
M SVB 2 32 -3.2561 2.8817
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 19 56
M SBL 1 1 21
M SMT 1 OCH3
M SVB 1 21 3.1947 3.949
S SKP 8
ID FL5FADGL0040
KNApSAcK_ID C00011130
NAME Isorhamnetin 3-O-beta-(6''-O-E-p-coumaroylglucoside)-7-O-beta-glucoside;7-(beta-D-Glucopyranosyloxy)-5-hydroxy-2-(4-hydroxy-3-methoxyphenyl)-3-[[6-O-[(2E)-3-(4-hydroxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl]oxy]-4H-1-benzopyran-4-one
CAS_RN 473919-26-5
FORMULA C37H38O19
EXACTMASS 786.200729034
AVERAGEMASS 786.68622
SMILES O[C@@H]([C@@H](O)6)[C@@H](OC(CO)[C@H](O)6)Oc(c5)cc(c1c5O)OC(c(c4)ccc(O)c4OC)=C(O[C@H](O2)[C@@H](O)[C@H]([C@H](C(COC(=O)C=Cc(c3)ccc(c3)O)2)O)O)C(=O)1
M END