Mol:FL5FACGS0070
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 44 48 0 0 0 0 0 0 0 0999 V2000 | + | 44 48 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.2548 2.8966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2548 2.8966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2548 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2548 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9693 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9693 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6838 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6838 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6838 2.8966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6838 2.8966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9693 3.3091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9693 3.3091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5404 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5404 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1741 2.0716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1741 2.0716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8886 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8886 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8886 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8886 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1741 0.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1741 0.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5404 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5404 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6031 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6031 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3175 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3175 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3175 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3175 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6031 0.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6031 0.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1741 -0.4034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1741 -0.4034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0320 2.0716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0320 2.0716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2548 0.4216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2548 0.4216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6031 -0.4034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6031 -0.4034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2757 3.2383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2757 3.2383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3123 1.7087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.3123 1.7087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1883 -1.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1883 -1.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5618 -1.8874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5618 -1.8874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0919 -1.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0919 -1.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8703 -0.9807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8703 -0.9807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1202 -0.6365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1202 -0.6365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5900 -1.2691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5900 -1.2691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0892 -0.8542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0892 -0.8542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4626 -0.9059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4626 -0.9059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6730 -1.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6730 -1.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2781 -2.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2781 -2.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5968 -1.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5968 -1.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8291 -2.1636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8291 -2.1636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4165 -2.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4165 -2.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6182 -2.6691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6182 -2.6691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8247 -2.8960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8247 -2.8960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2372 -2.1812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2372 -2.1812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.0356 -2.3903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.0356 -2.3903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1926 -1.8045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1926 -1.8045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6725 -1.5274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6725 -1.5274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3123 -2.6468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3123 -2.6468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8652 -2.6192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8652 -2.6192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7896 -3.3091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7896 -3.3091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 19 1 0 0 0 0 | + | 26 19 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0070 | + | ID FL5FACGS0070 |
| − | KNApSAcK_ID C00013842 | + | KNApSAcK_ID C00013842 |
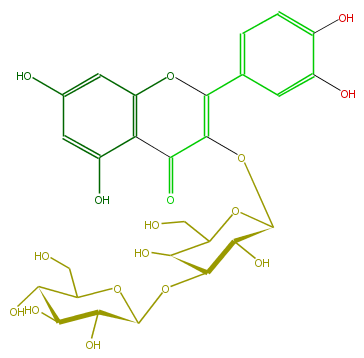
| − | NAME Quercetin 3-glucosyl-(1->3)-galactoside | + | NAME Quercetin 3-glucosyl-(1->3)-galactoside |
| − | CAS_RN - | + | CAS_RN - |
| − | FORMULA C27H30O17 | + | FORMULA C27H30O17 |
| − | EXACTMASS 626.148299534 | + | EXACTMASS 626.148299534 |
| − | AVERAGEMASS 626.5169000000001 | + | AVERAGEMASS 626.5169000000001 |
| − | SMILES c(c(O)5)cc(cc5O)C(=C1OC(O4)C(O)C(C(C4CO)O)OC(C(O)3)OC(C(O)C(O)3)CO)Oc(c2)c(c(O)cc2O)C1=O | + | SMILES c(c(O)5)cc(cc5O)C(=C1OC(O4)C(O)C(C(C4CO)O)OC(C(O)3)OC(C(O)C(O)3)CO)Oc(c2)c(c(O)cc2O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
44 48 0 0 0 0 0 0 0 0999 V2000
1.2548 2.8966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.2548 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9693 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6838 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6838 2.8966 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9693 3.3091 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5404 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1741 2.0716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.8886 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8886 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1741 0.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5404 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6031 2.0716 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3175 1.6591 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3175 0.8341 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6031 0.4216 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1741 -0.4034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.0320 2.0716 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2548 0.4216 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6031 -0.4034 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2757 3.2383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3123 1.7087 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.1883 -1.5433 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5618 -1.8874 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0919 -1.2549 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8703 -0.9807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1202 -0.6365 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5900 -1.2691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0892 -0.8542 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4626 -0.9059 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6730 -1.4610 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.2781 -2.1991 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5968 -1.6732 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8291 -2.1636 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.4165 -2.8783 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6182 -2.6691 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8247 -2.8960 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2372 -2.1812 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.0356 -2.3903 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1926 -1.8045 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6725 -1.5274 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3123 -2.6468 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8652 -2.6192 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7896 -3.3091 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 19 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
35 43 1 0 0 0 0
36 44 1 0 0 0 0
37 32 1 0 0 0 0
S SKP 8
ID FL5FACGS0070
KNApSAcK_ID C00013842
NAME Quercetin 3-glucosyl-(1->3)-galactoside
CAS_RN -
FORMULA C27H30O17
EXACTMASS 626.148299534
AVERAGEMASS 626.5169000000001
SMILES c(c(O)5)cc(cc5O)C(=C1OC(O4)C(O)C(C(C4CO)O)OC(C(O)3)OC(C(O)C(O)3)CO)Oc(c2)c(c(O)cc2O)C1=O
M END