Mol:FL5FACGL0037
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.0910 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0910 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.0910 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.0910 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3765 -0.2036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3765 -0.2036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3380 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3380 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3380 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3380 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3765 1.4465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3765 1.4465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0524 -0.2036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0524 -0.2036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7669 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7669 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7669 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7669 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0524 1.4465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0524 1.4465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0524 -0.8973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0524 -0.8973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4811 1.4463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4811 1.4463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2093 1.0259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2093 1.0259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9375 1.4463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9375 1.4463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9375 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9375 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2093 2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2093 2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4811 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4811 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3765 -0.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3765 -0.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8052 1.4463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8052 1.4463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6654 2.7074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6654 2.7074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5159 -0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5159 -0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6386 -0.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6386 -0.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8191 -0.6389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8191 -0.6389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6601 -0.9483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6601 -0.9483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2148 -1.6585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2148 -1.6585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0345 -1.2962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0345 -1.2962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1934 -0.9867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1934 -0.9867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9736 -0.8123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9736 -0.8123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5163 -1.7900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5163 -1.7900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2093 3.5482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2093 3.5482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5665 1.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5665 1.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0421 0.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0421 0.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2870 1.0570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2870 1.0570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5583 1.0649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5583 1.0649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0878 1.5946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0878 1.5946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7484 1.2459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7484 1.2459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2907 1.2914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2907 1.2914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7914 0.7873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7914 0.7873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6879 0.5975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6879 0.5975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9549 0.1046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9549 0.1046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8874 -1.8929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8874 -1.8929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.9834 -2.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9834 -2.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3016 -2.6901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3016 -2.6901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8260 -3.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8260 -3.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5811 -3.0886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5811 -3.0886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3098 -3.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3098 -3.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7803 -2.5511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7803 -2.5511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1197 -2.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1197 -2.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5994 -2.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5994 -2.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0767 -3.3583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0767 -3.3583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1802 -3.5482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1802 -3.5482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3508 1.7664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3508 1.7664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5163 1.7664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5163 1.7664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7681 2.7758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7681 2.7758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4788 -2.3407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4788 -2.3407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6376 -1.5311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6376 -1.5311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 1 19 1 0 0 0 0 | + | 1 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 8 21 1 0 0 0 0 | + | 8 21 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 26 29 1 0 0 0 0 | + | 26 29 1 0 0 0 0 |
| − | 16 30 1 0 0 0 0 | + | 16 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 19 1 0 0 0 0 | + | 34 19 1 0 0 0 0 |
| − | 23 21 1 0 0 0 0 | + | 23 21 1 0 0 0 0 |
| − | 22 40 1 0 0 0 0 | + | 22 40 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 41 25 1 0 0 0 0 | + | 41 25 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 42 1 0 0 0 0 | + | 46 42 1 0 0 0 0 |
| − | 52 53 2 0 0 0 0 | + | 52 53 2 0 0 0 0 |
| − | 52 54 1 0 0 0 0 | + | 52 54 1 0 0 0 0 |
| − | 36 52 1 0 0 0 0 | + | 36 52 1 0 0 0 0 |
| − | 48 55 1 0 0 0 0 | + | 48 55 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 52 53 54 | + | M SAL 1 3 52 53 54 |
| − | M SBL 1 1 59 | + | M SBL 1 1 59 |
| − | M SMT 1 ^ COOH | + | M SMT 1 ^ COOH |
| − | M SBV 1 59 0.6024 -0.5205 | + | M SBV 1 59 0.6024 -0.5205 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 55 56 | + | M SAL 2 2 55 56 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 60 0.6409 -0.5590 | + | M SBV 2 60 0.6409 -0.5590 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FACGL0037 | + | ID FL5FACGL0037 |
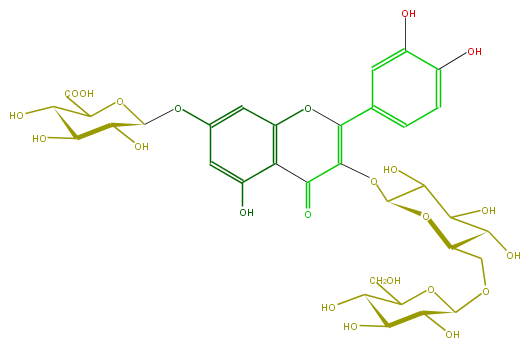
| − | FORMULA C33H38O23 | + | FORMULA C33H38O23 |
| − | EXACTMASS 802.180387522 | + | EXACTMASS 802.180387522 |
| − | AVERAGEMASS 802.64102 | + | AVERAGEMASS 802.64102 |
| − | SMILES O(C(C6O)OC(C(O)=O)C(O)C(O)6)c(c5)cc(c(c(O)5)4)OC(=C(C4=O)OC(C2O)OC(COC(O3)C(C(O)C(C(CO)3)O)O)C(O)C(O)2)c(c1)ccc(O)c(O)1 | + | SMILES O(C(C6O)OC(C(O)=O)C(O)C(O)6)c(c5)cc(c(c(O)5)4)OC(=C(C4=O)OC(C2O)OC(COC(O3)C(C(O)C(C(CO)3)O)O)C(O)C(O)2)c(c1)ccc(O)c(O)1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-1.0910 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.0910 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3765 -0.2036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3380 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3380 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3765 1.4465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0524 -0.2036 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7669 0.2090 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7669 1.0340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0524 1.4465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0524 -0.8973 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4811 1.4463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2093 1.0259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9375 1.4463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9375 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2093 2.7076 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4811 2.2871 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.3765 -0.8560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8052 1.4463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.6654 2.7074 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5159 -0.1560 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6386 -0.2765 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8191 -0.6389 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6601 -0.9483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2148 -1.6585 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0345 -1.2962 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.1934 -0.9867 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9736 -0.8123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5163 -1.7900 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.2093 3.5482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5665 1.4555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0421 0.7634 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2870 1.0570 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5583 1.0649 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0878 1.5946 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7484 1.2459 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.2907 1.2914 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7914 0.7873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6879 0.5975 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9549 0.1046 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8874 -1.8929 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.9834 -2.5912 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3016 -2.6901 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8260 -3.3822 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5811 -3.0886 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3098 -3.0807 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7803 -2.5511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1197 -2.8997 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5994 -2.9381 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0767 -3.3583 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1802 -3.5482 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.3508 1.7664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5163 1.7664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7681 2.7758 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4788 -2.3407 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6376 -1.5311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
1 19 1 0 0 0 0
15 20 1 0 0 0 0
8 21 1 0 0 0 0
22 23 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
27 28 1 0 0 0 0
26 29 1 0 0 0 0
16 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 19 1 0 0 0 0
23 21 1 0 0 0 0
22 40 1 0 0 0 0
41 42 1 0 0 0 0
41 25 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 42 1 0 0 0 0
52 53 2 0 0 0 0
52 54 1 0 0 0 0
36 52 1 0 0 0 0
48 55 1 0 0 0 0
55 56 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 52 53 54
M SBL 1 1 59
M SMT 1 ^ COOH
M SBV 1 59 0.6024 -0.5205
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 55 56
M SBL 2 1 60
M SMT 2 ^CH2OH
M SBV 2 60 0.6409 -0.5590
S SKP 5
ID FL5FACGL0037
FORMULA C33H38O23
EXACTMASS 802.180387522
AVERAGEMASS 802.64102
SMILES O(C(C6O)OC(C(O)=O)C(O)C(O)6)c(c5)cc(c(c(O)5)4)OC(=C(C4=O)OC(C2O)OC(COC(O3)C(C(O)C(C(CO)3)O)O)C(O)C(O)2)c(c1)ccc(O)c(O)1
M END