Mol:FL5FABGI0018
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 51 55 0 0 0 0 0 0 0 0999 V2000 | + | 51 55 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3382 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3382 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3382 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3382 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7819 -0.1394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7819 -0.1394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2256 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2256 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2256 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2256 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7819 1.1454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7819 1.1454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6693 -0.1394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6693 -0.1394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1130 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1130 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1130 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1130 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6693 1.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6693 1.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6693 -0.6402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6693 -0.6402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5875 1.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5875 1.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1544 0.9417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1544 0.9417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7214 1.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7214 1.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7214 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7214 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1544 2.2510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1544 2.2510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5875 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5875 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0027 1.2078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0027 1.2078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7819 -0.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7819 -0.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4709 -0.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4709 -0.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5236 -1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5236 -1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0590 -1.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0590 -1.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8891 -1.0255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8891 -1.0255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0590 -0.4275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0590 -0.4275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5236 -0.1183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5236 -0.1183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6935 -0.7128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6935 -0.7128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3500 -0.7128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3500 -0.7128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6611 -1.8241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6611 -1.8241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9299 -2.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9299 -2.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4177 -1.3307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4177 -1.3307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7819 1.7875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7819 1.7875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3380 2.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3380 2.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3380 2.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3380 2.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7831 3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.7831 3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8929 3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8929 3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.2900 -1.9632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.2900 -1.9632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0811 -2.4531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0811 -2.4531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6156 -2.2453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6156 -2.2453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1314 -2.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1314 -2.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7566 -1.8648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7566 -1.8648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2890 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2890 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8039 -2.2598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8039 -2.2598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4975 -2.4813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4975 -2.4813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9219 -2.7594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9219 -2.7594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0997 -2.7360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0997 -2.7360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9071 -3.0697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9071 -3.0697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7403 -2.7360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7403 -2.7360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2882 2.2509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2882 2.2509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0027 1.8384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0027 1.8384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.1664 -1.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.1664 -1.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6305 -1.4997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6305 -1.4997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 19 3 1 0 0 0 0 | + | 19 3 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 26 21 1 1 0 0 0 | + | 26 21 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 21 28 1 0 0 0 0 | + | 21 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 23 30 1 0 0 0 0 | + | 23 30 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 6 31 1 0 0 0 0 | + | 6 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 2 0 0 0 0 | + | 32 33 2 0 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 36 37 1 1 0 0 0 | + | 36 37 1 1 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 39 38 1 1 0 0 0 | + | 39 38 1 1 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 37 43 1 0 0 0 0 | + | 37 43 1 0 0 0 0 |
| − | 38 44 1 0 0 0 0 | + | 38 44 1 0 0 0 0 |
| − | 39 28 1 0 0 0 0 | + | 39 28 1 0 0 0 0 |
| − | 29 45 1 0 0 0 0 | + | 29 45 1 0 0 0 0 |
| − | 45 46 2 0 0 0 0 | + | 45 46 2 0 0 0 0 |
| − | 45 47 1 0 0 0 0 | + | 45 47 1 0 0 0 0 |
| − | 15 48 1 0 0 0 0 | + | 15 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 41 50 1 0 0 0 0 | + | 41 50 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 48 49 | + | M SAL 1 2 48 49 |
| − | M SBL 1 1 52 | + | M SBL 1 1 52 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 52 -4.2216 4.2864 | + | M SBV 1 52 -4.2216 4.2864 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 50 51 | + | M SAL 2 2 50 51 |
| − | M SBL 2 1 54 | + | M SBL 2 1 54 |
| − | M SMT 2 ^CH2OH | + | M SMT 2 ^CH2OH |
| − | M SBV 2 54 -4.9110 4.3576 | + | M SBV 2 54 -4.9110 4.3576 |
| − | S SKP 8 | + | S SKP 8 |
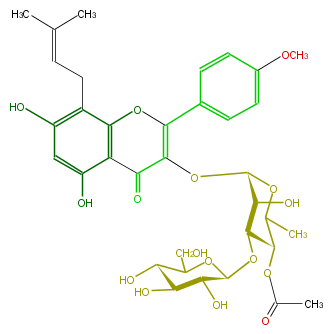
| − | ID FL5FABGI0018 | + | ID FL5FABGI0018 |
| − | KNApSAcK_ID C00006055 | + | KNApSAcK_ID C00006055 |
| − | NAME Epimedokoreanoside II | + | NAME Epimedokoreanoside II |
| − | CAS_RN 130756-12-6 | + | CAS_RN 130756-12-6 |
| − | FORMULA C35H42O16 | + | FORMULA C35H42O16 |
| − | EXACTMASS 718.247285296 | + | EXACTMASS 718.247285296 |
| − | AVERAGEMASS 718.69838 | + | AVERAGEMASS 718.69838 |
| − | SMILES O(C(C4O)OC(C)C(C(OC(O5)C(O)C(C(C5CO)O)O)4)OC(C)=O)C(=C2c(c3)ccc(OC)c3)C(=O)c(c1O)c(O2)c(c(c1)O)CC=C(C)C | + | SMILES O(C(C4O)OC(C)C(C(OC(O5)C(O)C(C(C5CO)O)O)4)OC(C)=O)C(=C2c(c3)ccc(OC)c3)C(=O)c(c1O)c(O2)c(c(c1)O)CC=C(C)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
51 55 0 0 0 0 0 0 0 0999 V2000
-2.3382 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3382 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7819 -0.1394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2256 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2256 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7819 1.1454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6693 -0.1394 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1130 0.1818 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1130 0.8242 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6693 1.1454 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6693 -0.6402 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.5875 1.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1544 0.9417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7214 1.2690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7214 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1544 2.2510 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5875 1.9237 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0027 1.2078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7819 -0.7021 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4709 -0.2071 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5236 -1.3108 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0590 -1.6199 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8891 -1.0255 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0590 -0.4275 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5236 -0.1183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6935 -0.7128 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3500 -0.7128 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6611 -1.8241 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.9299 -2.1020 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4177 -1.3307 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7819 1.7875 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3380 2.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3380 2.7493 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7831 3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8929 3.0697 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.2900 -1.9632 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0811 -2.4531 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6156 -2.2453 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1314 -2.2397 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7566 -1.8648 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.2890 -2.1116 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8039 -2.2598 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4975 -2.4813 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9219 -2.7594 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0997 -2.7360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9071 -3.0697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7403 -2.7360 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2882 2.2509 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0027 1.8384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.1664 -1.7132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6305 -1.4997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
4 3 1 0 0 0 0
1 18 1 0 0 0 0
19 3 1 0 0 0 0
8 20 1 0 0 0 0
22 21 1 1 0 0 0
22 23 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 26 1 1 0 0 0
26 21 1 1 0 0 0
26 27 1 0 0 0 0
21 28 1 0 0 0 0
22 29 1 0 0 0 0
23 30 1 0 0 0 0
25 20 1 0 0 0 0
6 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 2 0 0 0 0
33 34 1 0 0 0 0
33 35 1 0 0 0 0
36 37 1 1 0 0 0
37 38 1 1 0 0 0
39 38 1 1 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
41 36 1 0 0 0 0
36 42 1 0 0 0 0
37 43 1 0 0 0 0
38 44 1 0 0 0 0
39 28 1 0 0 0 0
29 45 1 0 0 0 0
45 46 2 0 0 0 0
45 47 1 0 0 0 0
15 48 1 0 0 0 0
48 49 1 0 0 0 0
41 50 1 0 0 0 0
50 51 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 48 49
M SBL 1 1 52
M SMT 1 OCH3
M SBV 1 52 -4.2216 4.2864
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 50 51
M SBL 2 1 54
M SMT 2 ^CH2OH
M SBV 2 54 -4.9110 4.3576
S SKP 8
ID FL5FABGI0018
KNApSAcK_ID C00006055
NAME Epimedokoreanoside II
CAS_RN 130756-12-6
FORMULA C35H42O16
EXACTMASS 718.247285296
AVERAGEMASS 718.69838
SMILES O(C(C4O)OC(C)C(C(OC(O5)C(O)C(C(C5CO)O)O)4)OC(C)=O)C(=C2c(c3)ccc(OC)c3)C(=O)c(c1O)c(O2)c(c(c1)O)CC=C(C)C
M END