Mol:FL5FAAGI0010
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 56 61 0 0 0 0 0 0 0 0999 V2000 | + | 56 61 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.8446 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8446 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8446 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8446 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1299 -0.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1299 -0.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4151 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4151 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4151 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4151 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1299 0.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1299 0.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2996 -0.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2996 -0.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0144 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0144 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0144 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0144 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2996 0.9466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2996 0.9466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3240 -1.4800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3240 -1.4800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7289 0.9465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7289 0.9465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4573 0.5259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4573 0.5259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1858 0.9465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1858 0.9465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1858 1.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1858 1.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4573 2.2083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4573 2.2083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7289 1.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7289 1.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5592 0.9465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5592 0.9465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9141 2.2082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9141 2.2082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7944 -0.7759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7944 -0.7759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1299 -1.5291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1299 -1.5291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1299 1.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1299 1.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8444 2.1842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8444 2.1842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8444 3.0093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8444 3.0093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1299 3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1299 3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5589 3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5589 3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.9813 1.0132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.9813 1.0132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5044 0.3836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5044 0.3836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8176 0.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8176 0.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1550 0.6578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1550 0.6578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.6364 1.1395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.6364 1.1395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2372 0.8224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2372 0.8224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5741 0.7895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5741 0.7895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2213 0.4496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2213 0.4496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1752 0.1538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1752 0.1538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7300 -2.4149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7300 -2.4149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1378 -2.9118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1378 -2.9118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9089 -2.9664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9089 -2.9664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5390 -3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5390 -3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1312 -2.9249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1312 -2.9249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3600 -2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3600 -2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0556 -2.3537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0556 -2.3537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0053 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0053 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.5741 -3.3424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.5741 -3.3424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0909 -1.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0909 -1.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0909 -2.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0909 -2.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5480 -2.0888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5480 -2.0888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1892 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1892 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1892 -1.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1892 -1.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7320 -1.7423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7320 -1.7423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4886 -2.8675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4886 -2.8675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6069 -2.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6069 -2.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7016 -2.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7016 -2.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5341 -2.5816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.5341 -2.5816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8370 1.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8370 1.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1996 2.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1996 2.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 15 19 1 0 0 0 0 | + | 15 19 1 0 0 0 0 |
| − | 8 20 1 0 0 0 0 | + | 8 20 1 0 0 0 0 |
| − | 3 21 1 0 0 0 0 | + | 3 21 1 0 0 0 0 |
| − | 6 22 1 0 0 0 0 | + | 6 22 1 0 0 0 0 |
| − | 22 23 1 0 0 0 0 | + | 22 23 1 0 0 0 0 |
| − | 23 24 2 0 0 0 0 | + | 23 24 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 27 28 1 1 0 0 0 | + | 27 28 1 1 0 0 0 |
| − | 28 29 1 1 0 0 0 | + | 28 29 1 1 0 0 0 |
| − | 30 29 1 1 0 0 0 | + | 30 29 1 1 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 27 1 0 0 0 0 | + | 32 27 1 0 0 0 0 |
| − | 27 33 1 0 0 0 0 | + | 27 33 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 29 35 1 0 0 0 0 | + | 29 35 1 0 0 0 0 |
| − | 30 18 1 0 0 0 0 | + | 30 18 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 1 1 0 0 0 | + | 37 38 1 1 0 0 0 |
| − | 38 39 1 1 0 0 0 | + | 38 39 1 1 0 0 0 |
| − | 40 39 1 1 0 0 0 | + | 40 39 1 1 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 36 1 0 0 0 0 | + | 41 36 1 0 0 0 0 |
| − | 36 42 1 0 0 0 0 | + | 36 42 1 0 0 0 0 |
| − | 41 43 1 0 0 0 0 | + | 41 43 1 0 0 0 0 |
| − | 40 44 1 0 0 0 0 | + | 40 44 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 47 48 1 1 0 0 0 | + | 47 48 1 1 0 0 0 |
| − | 49 48 1 1 0 0 0 | + | 49 48 1 1 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 47 51 1 0 0 0 0 | + | 47 51 1 0 0 0 0 |
| − | 46 52 1 0 0 0 0 | + | 46 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 48 54 1 0 0 0 0 | + | 48 54 1 0 0 0 0 |
| − | 49 20 1 0 0 0 0 | + | 49 20 1 0 0 0 0 |
| − | 37 54 1 0 0 0 0 | + | 37 54 1 0 0 0 0 |
| − | 55 56 1 0 0 0 0 | + | 55 56 1 0 0 0 0 |
| − | 32 55 1 0 0 0 0 | + | 32 55 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 55 56 | + | M SAL 1 2 55 56 |
| − | M SBL 1 1 61 | + | M SBL 1 1 61 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 61 0.5998 -0.5510 | + | M SBV 1 61 0.5998 -0.5510 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FAAGI0010 | + | ID FL5FAAGI0010 |
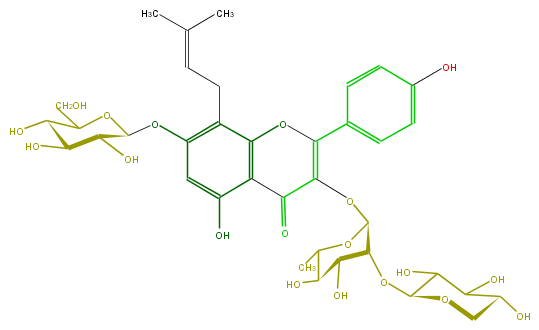
| − | FORMULA C37H46O19 | + | FORMULA C37H46O19 |
| − | EXACTMASS 794.26332929 | + | EXACTMASS 794.26332929 |
| − | AVERAGEMASS 794.74974 | + | AVERAGEMASS 794.74974 |
| − | SMILES Oc(c6)ccc(c6)C(=C(OC(C4OC(O5)C(C(O)C(O)C5)O)OC(C)C(C4O)O)1)Oc(c(CC=C(C)C)2)c(c(cc2OC(O3)C(C(O)C(C3CO)O)O)O)C1=O | + | SMILES Oc(c6)ccc(c6)C(=C(OC(C4OC(O5)C(C(O)C(O)C5)O)OC(C)C(C4O)O)1)Oc(c(CC=C(C)C)2)c(c(cc2OC(O3)C(C(O)C(C3CO)O)O)O)C1=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
56 61 0 0 0 0 0 0 0 0999 V2000
-1.8446 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8446 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1299 -0.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4151 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4151 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1299 0.9466 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2996 -0.7041 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0144 -0.2914 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0144 0.5340 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2996 0.9466 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3240 -1.4800 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7289 0.9465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4573 0.5259 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1858 0.9465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1858 1.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4573 2.2083 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7289 1.7877 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5592 0.9465 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9141 2.2082 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7944 -0.7759 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1299 -1.5291 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1299 1.7717 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8444 2.1842 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8444 3.0093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1299 3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5589 3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.9813 1.0132 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5044 0.3836 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8176 0.6507 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.1550 0.6578 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.6364 1.1395 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.2372 0.8224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5741 0.7895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2213 0.4496 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1752 0.1538 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7300 -2.4149 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1378 -2.9118 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9089 -2.9664 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.5390 -3.4217 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1312 -2.9249 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.3600 -2.8702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0556 -2.3537 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0053 -2.5235 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.5741 -3.3424 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0909 -1.9040 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0909 -2.5614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5480 -2.0888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1892 -1.9273 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1892 -1.2698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7320 -1.7423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4886 -2.8675 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.6069 -2.6280 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.7016 -2.2599 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5341 -2.5816 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.8370 1.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1996 2.3695 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
15 19 1 0 0 0 0
8 20 1 0 0 0 0
3 21 1 0 0 0 0
6 22 1 0 0 0 0
22 23 1 0 0 0 0
23 24 2 0 0 0 0
24 25 1 0 0 0 0
24 26 1 0 0 0 0
27 28 1 1 0 0 0
28 29 1 1 0 0 0
30 29 1 1 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
32 27 1 0 0 0 0
27 33 1 0 0 0 0
28 34 1 0 0 0 0
29 35 1 0 0 0 0
30 18 1 0 0 0 0
36 37 1 0 0 0 0
37 38 1 1 0 0 0
38 39 1 1 0 0 0
40 39 1 1 0 0 0
40 41 1 0 0 0 0
41 36 1 0 0 0 0
36 42 1 0 0 0 0
41 43 1 0 0 0 0
40 44 1 0 0 0 0
45 46 1 0 0 0 0
46 47 1 1 0 0 0
47 48 1 1 0 0 0
49 48 1 1 0 0 0
49 50 1 0 0 0 0
50 45 1 0 0 0 0
47 51 1 0 0 0 0
46 52 1 0 0 0 0
45 53 1 0 0 0 0
48 54 1 0 0 0 0
49 20 1 0 0 0 0
37 54 1 0 0 0 0
55 56 1 0 0 0 0
32 55 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 55 56
M SBL 1 1 61
M SMT 1 ^ CH2OH
M SBV 1 61 0.5998 -0.5510
S SKP 5
ID FL5FAAGI0010
FORMULA C37H46O19
EXACTMASS 794.26332929
AVERAGEMASS 794.74974
SMILES Oc(c6)ccc(c6)C(=C(OC(C4OC(O5)C(C(O)C(O)C5)O)OC(C)C(C4O)O)1)Oc(c(CC=C(C)C)2)c(c(cc2OC(O3)C(C(O)C(C3CO)O)O)O)C1=O
M END