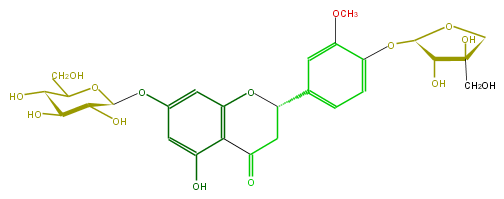
Mol:FL2FCDGS0001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 46 0 0 0 0 0 0 0 0999 V2000 | + | 42 46 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.1002 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1002 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3857 -2.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3857 -2.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6712 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6712 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6712 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6712 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3857 -0.4153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3857 -0.4153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1002 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1002 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0433 -2.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0433 -2.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7576 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7576 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7576 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7576 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0433 -0.4153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0433 -0.4153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0433 -2.7839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0433 -2.7839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3857 -2.8895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3857 -2.8895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7953 -0.4264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7953 -0.4264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5528 -0.4171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5528 -0.4171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2781 -0.8357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2781 -0.8357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0033 -0.4171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0033 -0.4171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0033 0.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0033 0.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2781 0.8391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2781 0.8391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5528 0.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5528 0.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7270 0.8014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7270 0.8014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.3640 -0.4193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.3640 -0.4193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.8920 -1.0424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.8920 -1.0424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2123 -0.7781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2123 -0.7781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.5564 -0.7710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.5564 -0.7710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0331 -0.2943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0331 -0.2943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7274 -0.5436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7274 -0.5436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.9902 -0.5326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.9902 -0.5326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5218 -0.9908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5218 -0.9908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4709 -1.1857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4709 -1.1857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.3420 0.9012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.3420 0.9012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8628 0.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8628 0.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6798 0.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6798 0.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.1912 0.9892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.1912 0.9892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2338 1.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2338 1.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8706 -0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8706 -0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.6564 0.9542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.6564 0.9542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2781 1.6470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2781 1.6470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8593 2.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8593 2.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7105 -0.2513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7105 -0.2513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5463 -0.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5463 -0.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.2189 0.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.2189 0.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -6.5463 0.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -6.5463 0.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 3 7 1 0 0 0 0 | + | 3 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 2 12 1 0 0 0 0 | + | 2 12 1 0 0 0 0 |
| − | 6 13 1 0 0 0 0 | + | 6 13 1 0 0 0 0 |
| − | 9 14 1 6 0 0 0 | + | 9 14 1 6 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 18 1 0 0 0 0 | + | 17 18 1 0 0 0 0 |
| − | 18 19 2 0 0 0 0 | + | 18 19 2 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 17 20 1 0 0 0 0 | + | 17 20 1 0 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 24 23 1 1 0 0 0 | + | 24 23 1 1 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 13 1 0 0 0 0 | + | 24 13 1 0 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 33 32 1 1 0 0 0 | + | 33 32 1 1 0 0 0 |
| − | 33 34 1 0 0 0 0 | + | 33 34 1 0 0 0 0 |
| − | 34 30 1 0 0 0 0 | + | 34 30 1 0 0 0 0 |
| − | 31 35 1 0 0 0 0 | + | 31 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 30 20 1 0 0 0 0 | + | 30 20 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 18 37 1 0 0 0 0 | + | 18 37 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 32 39 1 0 0 0 0 | + | 32 39 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 37 38 | + | M SAL 1 2 37 38 |
| − | M SBL 1 1 42 | + | M SBL 1 1 42 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SBV 1 42 0.0000 -0.8079 | + | M SBV 1 42 0.0000 -0.8079 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 39 40 | + | M SAL 2 2 39 40 |
| − | M SBL 2 1 44 | + | M SBL 2 1 44 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SBV 2 44 -0.0307 0.6829 | + | M SBV 2 44 -0.0307 0.6829 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 41 42 | + | M SAL 3 2 41 42 |
| − | M SBL 3 1 46 | + | M SBL 3 1 46 |
| − | M SMT 3 ^ CH2OH | + | M SMT 3 ^ CH2OH |
| − | M SBV 3 46 0.4915 -0.5597 | + | M SBV 3 46 0.4915 -0.5597 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL2FCDGS0001 | + | ID FL2FCDGS0001 |
| − | FORMULA C27H32O15 | + | FORMULA C27H32O15 |
| − | EXACTMASS 596.174120354 | + | EXACTMASS 596.174120354 |
| − | AVERAGEMASS 596.5339799999999 | + | AVERAGEMASS 596.5339799999999 |
| − | SMILES c(c4)(OC(C(O)5)OCC(CO)5O)c(cc(c4)C(C1)Oc(c2)c(c(O)cc(OC(C(O)3)OC(CO)C(O)C3O)2)C(=O)1)OC | + | SMILES c(c4)(OC(C(O)5)OCC(CO)5O)c(cc(c4)C(C1)Oc(c2)c(c(O)cc(OC(C(O)3)OC(CO)C(O)C3O)2)C(=O)1)OC |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 46 0 0 0 0 0 0 0 0999 V2000
-2.1002 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3857 -2.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6712 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6712 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.3857 -0.4153 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1002 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0433 -2.0652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7576 -1.6528 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7576 -0.8277 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0433 -0.4153 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0433 -2.7839 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3857 -2.8895 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7953 -0.4264 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5528 -0.4171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2781 -0.8357 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0033 -0.4171 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0033 0.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2781 0.8391 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5528 0.4203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7270 0.8014 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.3640 -0.4193 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.8920 -1.0424 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2123 -0.7781 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.5564 -0.7710 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.0331 -0.2943 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7274 -0.5436 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.9902 -0.5326 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.5218 -0.9908 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4709 -1.1857 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.3420 0.9012 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.8628 0.4288 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.6798 0.4315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.1912 0.9892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2338 1.3119 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8706 -0.1795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.6564 0.9542 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2781 1.6470 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8593 2.8895 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7105 -0.2513 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5463 -0.1041 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.2189 0.0161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-6.5463 0.3619 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
3 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 1 0 0 0 0
10 4 1 0 0 0 0
7 11 2 0 0 0 0
2 12 1 0 0 0 0
6 13 1 0 0 0 0
9 14 1 6 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 18 1 0 0 0 0
18 19 2 0 0 0 0
19 14 1 0 0 0 0
17 20 1 0 0 0 0
21 22 1 1 0 0 0
22 23 1 1 0 0 0
24 23 1 1 0 0 0
24 25 1 0 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 13 1 0 0 0 0
30 31 1 1 0 0 0
31 32 1 1 0 0 0
33 32 1 1 0 0 0
33 34 1 0 0 0 0
34 30 1 0 0 0 0
31 35 1 0 0 0 0
32 36 1 0 0 0 0
30 20 1 0 0 0 0
37 38 1 0 0 0 0
18 37 1 0 0 0 0
39 40 1 0 0 0 0
32 39 1 0 0 0 0
41 42 1 0 0 0 0
26 41 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 37 38
M SBL 1 1 42
M SMT 1 OCH3
M SBV 1 42 0.0000 -0.8079
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 39 40
M SBL 2 1 44
M SMT 2 CH2OH
M SBV 2 44 -0.0307 0.6829
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 41 42
M SBL 3 1 46
M SMT 3 ^ CH2OH
M SBV 3 46 0.4915 -0.5597
S SKP 5
ID FL2FCDGS0001
FORMULA C27H32O15
EXACTMASS 596.174120354
AVERAGEMASS 596.5339799999999
SMILES c(c4)(OC(C(O)5)OCC(CO)5O)c(cc(c4)C(C1)Oc(c2)c(c(O)cc(OC(C(O)3)OC(CO)C(O)C3O)2)C(=O)1)OC
M END