Mol:BMMCBZ3Sa008
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 58 61 0 0 1 0 0 0 0 0999 V2000 | + | 58 61 0 0 1 0 0 0 0 0999 V2000 |
| − | 4.9344 -3.3593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.9344 -3.3593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6254 -4.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6254 -4.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6473 -4.5183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6473 -4.5183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9781 -3.7751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9781 -3.7751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.2872 -2.8241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.2872 -2.8241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2653 -2.6161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2653 -2.6161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.9126 -3.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.9126 -3.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.2216 -4.5183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.2216 -4.5183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2946 -5.0535 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2946 -5.0535 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0000 -3.9830 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0000 -3.9830 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.2697 -2.5373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.2697 -2.5373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.4037 -2.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.4037 -2.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.4037 -1.0373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.4037 -1.0373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.2697 -0.5373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.2697 -0.5373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.1357 -1.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.1357 -1.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.1357 -2.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.1357 -2.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.8789 -0.3681 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.8789 -0.3681 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 24.4721 0.5454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 24.4721 0.5454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.4776 0.4409 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.4776 0.4409 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 25.0018 -2.5373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 25.0018 -2.5373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.8085 1.1840 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 22.8085 1.1840 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 23.0164 2.1622 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 23.0164 2.1622 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 22.1504 2.6622 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 22.1504 2.6622 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 21.4072 1.9930 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 21.4072 1.9930 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 20.4291 2.2010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 20.4291 2.2010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.9299 2.5689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.9299 2.5689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.0458 3.6567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.0458 3.6567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 21.8140 1.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 21.8140 1.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 19.7599 1.4578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 19.7599 1.4578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.8549 4.2445 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.8549 4.2445 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.4426 3.4355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.4426 3.4355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 22.2671 5.0535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 22.2671 5.0535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 23.6639 4.8323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 23.6639 4.8323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.7818 1.6657 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.7818 1.6657 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.5739 0.6876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.5739 0.6876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 18.9897 2.6439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 18.9897 2.6439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.8037 1.8736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.8037 1.8736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.1345 1.1305 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.1345 1.1305 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 17.8777 0.4614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 17.8777 0.4614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.3914 1.7996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.3914 1.7996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 16.4654 0.3873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 16.4654 0.3873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.4872 0.5953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.4872 0.5953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.8181 -0.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.8181 -0.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 15.5613 -0.8170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 15.5613 -0.8170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.0750 0.5212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.0750 0.5212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.1490 -0.8910 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | 14.1490 -0.8910 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | 13.1708 -0.6831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 13.1708 -0.6831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.5017 -1.4263 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.5017 -1.4263 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 11.5236 -1.2184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 11.5236 -1.2184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 10.8544 -1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 10.8544 -1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.8763 -1.7536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.8763 -1.7536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.2071 -2.4967 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.2071 -2.4967 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 8.2290 -2.2888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 8.2290 -2.2888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 7.5599 -3.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 7.5599 -3.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 6.5817 -2.8241 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | 6.5817 -2.8241 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 14.4580 -1.8421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 14.4580 -1.8421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 12.8618 0.2679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 12.8618 0.2679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 9.5673 -0.8025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 9.5673 -0.8025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4 10 1 0 0 0 0 | + | 4 10 1 0 0 0 0 |
| − | 2 1 2 0 0 0 0 | + | 2 1 2 0 0 0 0 |
| − | 1 6 1 0 0 0 0 | + | 1 6 1 0 0 0 0 |
| − | 1 7 1 0 0 0 0 | + | 1 7 1 0 0 0 0 |
| − | 6 5 2 0 0 0 0 | + | 6 5 2 0 0 0 0 |
| − | 7 8 2 0 0 0 0 | + | 7 8 2 0 0 0 0 |
| − | 5 4 1 0 0 0 0 | + | 5 4 1 0 0 0 0 |
| − | 7 55 1 0 0 0 0 | + | 7 55 1 0 0 0 0 |
| − | 4 3 2 0 0 0 0 | + | 4 3 2 0 0 0 0 |
| − | 2 9 1 0 0 0 0 | + | 2 9 1 0 0 0 0 |
| − | 3 2 1 0 0 0 0 | + | 3 2 1 0 0 0 0 |
| − | 16 15 1 0 0 0 0 | + | 16 15 1 0 0 0 0 |
| − | 34 36 1 0 0 0 0 | + | 34 36 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 34 35 2 0 0 0 0 | + | 34 35 2 0 0 0 0 |
| − | 38 40 2 0 0 0 0 | + | 38 40 2 0 0 0 0 |
| − | 34 37 1 0 0 0 0 | + | 34 37 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 22 26 1 1 0 0 0 | + | 22 26 1 1 0 0 0 |
| − | 38 41 1 0 0 0 0 | + | 38 41 1 0 0 0 0 |
| − | 23 27 1 1 0 0 0 | + | 23 27 1 1 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 42 43 1 0 0 0 0 | + | 42 43 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 43 46 1 0 0 0 0 | + | 43 46 1 0 0 0 0 |
| − | 30 33 1 0 0 0 0 | + | 30 33 1 0 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 27 30 1 0 0 0 0 | + | 27 30 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 21 19 1 0 0 0 0 | + | 21 19 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 24 28 1 6 0 0 0 | + | 24 28 1 6 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 19 14 1 0 0 0 0 | + | 19 14 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 15 17 1 0 0 0 0 | + | 15 17 1 0 0 0 0 |
| − | 53 54 1 0 0 0 0 | + | 53 54 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 18 19 1 0 0 0 0 | + | 18 19 1 0 0 0 0 |
| − | 43 45 1 0 0 0 0 | + | 43 45 1 0 0 0 0 |
| − | 21 28 1 6 0 0 0 | + | 21 28 1 6 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 23 22 1 0 0 0 0 | + | 23 22 1 0 0 0 0 |
| − | 46 56 1 1 0 0 0 | + | 46 56 1 1 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 47 57 2 0 0 0 0 | + | 47 57 2 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 51 58 2 0 0 0 0 | + | 51 58 2 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 29 34 1 0 0 0 0 | + | 29 34 1 0 0 0 0 |
| − | 15 14 2 0 0 0 0 | + | 15 14 2 0 0 0 0 |
| − | 14 13 1 0 0 0 0 | + | 14 13 1 0 0 0 0 |
| − | 13 12 2 0 0 0 0 | + | 13 12 2 0 0 0 0 |
| − | 12 11 1 0 0 0 0 | + | 12 11 1 0 0 0 0 |
| − | 11 16 2 0 0 0 0 | + | 11 16 2 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 30 32 2 0 0 0 0 | + | 30 32 2 0 0 0 0 |
| − | S SKP 7 | + | S SKP 7 |
| − | ID BMMCBZ3Sa008 | + | ID BMMCBZ3Sa008 |
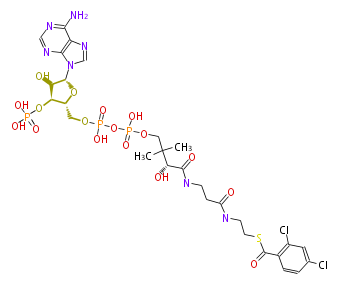
| − | NAME 2,4-Dichloro-benzoyl-CoA | + | NAME 2,4-Dichloro-benzoyl-CoA |
| − | FORMULA C28H38Cl2N7O17P3S | + | FORMULA C28H38Cl2N7O17P3S |
| − | EXACTMASS 939.0634 | + | EXACTMASS 939.0634 |
| − | AVERAGEMASS 940.5309 | + | AVERAGEMASS 940.5309 |
| − | SMILES c(c1Cl)cc(C(SCCNC(CCNC(=O)[C@@H](C(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)2)O[C@@H](n(c34)cnc3c(ncn4)N)[C@@H]2O)(O)=O)(O)=O)O)=O)=O)c(Cl)c1 | + | SMILES c(c1Cl)cc(C(SCCNC(CCNC(=O)[C@@H](C(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)2)O[C@@H](n(c34)cnc3c(ncn4)N)[C@@H]2O)(O)=O)(O)=O)O)=O)=O)c(Cl)c1 |
| − | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06671 | + | KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06671 |
M END | M END | ||
| − | |||
Revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
58 61 0 0 1 0 0 0 0 0999 V2000
4.9344 -3.3593 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6254 -4.3103 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6473 -4.5183 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9781 -3.7751 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.2872 -2.8241 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.2653 -2.6161 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.9126 -3.5672 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.2216 -4.5183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.2946 -5.0535 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0
2.0000 -3.9830 0.0000 Cl 0 0 0 0 0 0 0 0 0 0 0 0
23.2697 -2.5373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.4037 -2.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
22.4037 -1.0373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
23.2697 -0.5373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.1357 -1.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.1357 -2.0373 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
24.8789 -0.3681 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
24.4721 0.5454 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
23.4776 0.4409 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
25.0018 -2.5373 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
22.8085 1.1840 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
23.0164 2.1622 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
22.1504 2.6622 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
21.4072 1.9930 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
20.4291 2.2010 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
23.9299 2.5689 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.0458 3.6567 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
21.8140 1.0795 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
19.7599 1.4578 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.8549 4.2445 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
23.4426 3.4355 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
22.2671 5.0535 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
23.6639 4.8323 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
18.7818 1.6657 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
18.5739 0.6876 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
18.9897 2.6439 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17.8037 1.8736 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
17.1345 1.1305 0.0000 P 0 0 0 0 0 0 0 0 0 0 0 0
17.8777 0.4614 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.3914 1.7996 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
16.4654 0.3873 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
15.4872 0.5953 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.8181 -0.1479 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
15.5613 -0.8170 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.0750 0.5212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
14.1490 -0.8910 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
13.1708 -0.6831 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
12.5017 -1.4263 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
11.5236 -1.2184 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
10.8544 -1.9615 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.8763 -1.7536 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
9.2071 -2.4967 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
8.2290 -2.2888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.5599 -3.0320 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5817 -2.8241 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
14.4580 -1.8421 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
12.8618 0.2679 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
9.5673 -0.8025 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4 10 1 0 0 0 0
2 1 2 0 0 0 0
1 6 1 0 0 0 0
1 7 1 0 0 0 0
6 5 2 0 0 0 0
7 8 2 0 0 0 0
5 4 1 0 0 0 0
7 55 1 0 0 0 0
4 3 2 0 0 0 0
2 9 1 0 0 0 0
3 2 1 0 0 0 0
16 15 1 0 0 0 0
34 36 1 0 0 0 0
37 38 1 0 0 0 0
34 35 2 0 0 0 0
38 40 2 0 0 0 0
34 37 1 0 0 0 0
38 39 1 0 0 0 0
22 26 1 1 0 0 0
38 41 1 0 0 0 0
23 27 1 1 0 0 0
41 42 1 0 0 0 0
42 43 1 0 0 0 0
30 31 1 0 0 0 0
43 46 1 0 0 0 0
30 33 1 0 0 0 0
46 47 1 0 0 0 0
27 30 1 0 0 0 0
47 48 1 0 0 0 0
48 49 1 0 0 0 0
21 19 1 0 0 0 0
49 50 1 0 0 0 0
24 28 1 6 0 0 0
50 51 1 0 0 0 0
23 24 1 0 0 0 0
51 52 1 0 0 0 0
19 14 1 0 0 0 0
52 53 1 0 0 0 0
15 17 1 0 0 0 0
53 54 1 0 0 0 0
17 18 2 0 0 0 0
54 55 1 0 0 0 0
18 19 1 0 0 0 0
43 45 1 0 0 0 0
21 28 1 6 0 0 0
43 44 1 0 0 0 0
23 22 1 0 0 0 0
46 56 1 1 0 0 0
21 22 1 0 0 0 0
47 57 2 0 0 0 0
24 25 1 0 0 0 0
51 58 2 0 0 0 0
25 29 1 0 0 0 0
29 34 1 0 0 0 0
15 14 2 0 0 0 0
14 13 1 0 0 0 0
13 12 2 0 0 0 0
12 11 1 0 0 0 0
11 16 2 0 0 0 0
16 20 1 0 0 0 0
30 32 2 0 0 0 0
S SKP 7
ID BMMCBZ3Sa008
NAME 2,4-Dichloro-benzoyl-CoA
FORMULA C28H38Cl2N7O17P3S
EXACTMASS 939.0634
AVERAGEMASS 940.5309
SMILES c(c1Cl)cc(C(SCCNC(CCNC(=O)[C@@H](C(C)(C)COP(OP(OC[C@H]([C@@H](OP(O)(O)=O)2)O[C@@H](n(c34)cnc3c(ncn4)N)[C@@H]2O)(O)=O)(O)=O)O)=O)=O)c(Cl)c1
KEGG http://www.genome.jp/dbget-bin/www_bget?compound+C06671
M END