Mol:FLNACCGS0003
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 36 39 0 0 0 0 0 0 0 0999 V2000 | + | 36 39 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.5720 1.4802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5720 1.4802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0157 1.8014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0157 1.8014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5404 1.4804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5404 1.4804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5404 0.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5404 0.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1171 0.4814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1171 0.4814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6938 0.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6938 0.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6938 1.4804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6938 1.4804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1171 1.8133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1171 1.8133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5720 0.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5720 0.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0157 0.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0157 0.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1171 -0.1841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1171 -0.1841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5642 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5642 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5642 -1.1418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5642 -1.1418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1171 -1.4610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1171 -1.4610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6700 -1.1418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6700 -1.1418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6700 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6700 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2702 1.8131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2702 1.8131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1171 -2.0990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1171 -2.0990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0134 -0.1343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0134 -0.1343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.7924 -0.4516 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -1.7924 -0.4516 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -1.4342 -0.9245 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -1.4342 -0.9245 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.9184 -0.7239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.9184 -0.7239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.4207 -0.7185 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -0.4207 -0.7185 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -0.7824 -0.3568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7824 -0.3568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3093 -0.5460 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -1.3093 -0.5460 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -2.1574 -0.3188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1574 -0.3188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9926 -0.9517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9926 -0.9517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6229 -1.2200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6229 -1.2200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6332 0.1486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6332 0.1486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8412 0.5948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8412 0.5948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4322 0.6465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4322 0.6465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7284 0.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7284 0.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6297 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6297 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2226 -1.4608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2226 -1.4608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.9293 2.0990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.9293 2.0990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4293 2.9650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4293 2.9650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 7 1 0 0 0 0 | + | 6 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 3 1 0 0 0 0 | + | 8 3 1 0 0 0 0 |
| − | 1 9 1 0 0 0 0 | + | 1 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 4 1 0 0 0 0 | + | 10 4 1 0 0 0 0 |
| − | 5 11 1 0 0 0 0 | + | 5 11 1 0 0 0 0 |
| − | 11 12 2 0 0 0 0 | + | 11 12 2 0 0 0 0 |
| − | 12 13 1 0 0 0 0 | + | 12 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 11 1 0 0 0 0 | + | 16 11 1 0 0 0 0 |
| − | 7 17 2 0 0 0 0 | + | 7 17 2 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 10 19 1 0 0 0 0 | + | 10 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 25 29 1 0 0 0 0 | + | 25 29 1 0 0 0 0 |
| − | 23 19 1 0 0 0 0 | + | 23 19 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 2 0 0 0 0 | + | 31 32 2 0 0 0 0 |
| − | 31 33 1 0 0 0 0 | + | 31 33 1 0 0 0 0 |
| − | 15 34 1 0 0 0 0 | + | 15 34 1 0 0 0 0 |
| − | 1 35 1 0 0 0 0 | + | 1 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 35 36 | + | M SAL 1 2 35 36 |
| − | M SBL 1 1 38 | + | M SBL 1 1 38 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 38 -0.9293 2.099 | + | M SVB 1 38 -0.9293 2.099 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FLNACCGS0003 | + | ID FLNACCGS0003 |
| − | KNApSAcK_ID C00010247 | + | KNApSAcK_ID C00010247 |
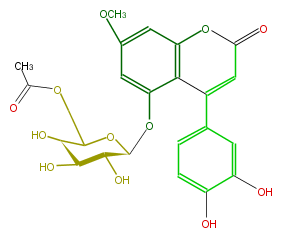
| − | NAME 5,3',4'-Trihydroxy-7-methoxy-4-phenylcoumarin 5-O-(6''-acetyl)-galactoside | + | NAME 5,3',4'-Trihydroxy-7-methoxy-4-phenylcoumarin 5-O-(6''-acetyl)-galactoside |
| − | CAS_RN 117783-71-8 | + | CAS_RN 117783-71-8 |
| − | FORMULA C24H24O12 | + | FORMULA C24H24O12 |
| − | EXACTMASS 504.126776232 | + | EXACTMASS 504.126776232 |
| − | AVERAGEMASS 504.44016 | + | AVERAGEMASS 504.44016 |
| − | SMILES O(c(c1)cc(O[C@@H]([C@H]4O)OC(COC(C)=O)[C@@H]([C@H](O)4)O)c(C(c(c3)cc(c(c3)O)O)=2)c(OC(=O)C2)1)C | + | SMILES O(c(c1)cc(O[C@@H]([C@H]4O)OC(COC(C)=O)[C@@H]([C@H](O)4)O)c(C(c(c3)cc(c(c3)O)O)=2)c(OC(=O)C2)1)C |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
36 39 0 0 0 0 0 0 0 0999 V2000
-0.5720 1.4802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0157 1.8014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5404 1.4804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5404 0.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1171 0.4814 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6938 0.8144 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6938 1.4804 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1171 1.8133 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5720 0.8382 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0157 0.5172 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1171 -0.1841 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5642 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5642 -1.1418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1171 -1.4610 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6700 -1.1418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6700 -0.5033 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2702 1.8131 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.1171 -2.0990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0134 -0.1343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.7924 -0.4516 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-1.4342 -0.9245 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.9184 -0.7239 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.4207 -0.7185 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-0.7824 -0.3568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3093 -0.5460 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-2.1574 -0.3188 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9926 -0.9517 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.6229 -1.2200 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.6332 0.1486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8412 0.5948 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4322 0.6465 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.7284 0.2236 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6297 1.0702 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2226 -1.4608 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.9293 2.0990 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4293 2.9650 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 7 1 0 0 0 0
7 8 1 0 0 0 0
8 3 1 0 0 0 0
1 9 1 0 0 0 0
9 10 2 0 0 0 0
10 4 1 0 0 0 0
5 11 1 0 0 0 0
11 12 2 0 0 0 0
12 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 11 1 0 0 0 0
7 17 2 0 0 0 0
14 18 1 0 0 0 0
10 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
22 28 1 0 0 0 0
25 29 1 0 0 0 0
23 19 1 0 0 0 0
29 30 1 0 0 0 0
30 31 1 0 0 0 0
31 32 2 0 0 0 0
31 33 1 0 0 0 0
15 34 1 0 0 0 0
1 35 1 0 0 0 0
35 36 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 35 36
M SBL 1 1 38
M SMT 1 OCH3
M SVB 1 38 -0.9293 2.099
S SKP 8
ID FLNACCGS0003
KNApSAcK_ID C00010247
NAME 5,3',4'-Trihydroxy-7-methoxy-4-phenylcoumarin 5-O-(6''-acetyl)-galactoside
CAS_RN 117783-71-8
FORMULA C24H24O12
EXACTMASS 504.126776232
AVERAGEMASS 504.44016
SMILES O(c(c1)cc(O[C@@H]([C@H]4O)OC(COC(C)=O)[C@@H]([C@H](O)4)O)c(C(c(c3)cc(c(c3)O)O)=2)c(OC(=O)C2)1)C
M END