Mol:FL5FFCGL0007
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 46 50 0 0 0 0 0 0 0 0999 V2000 | + | 46 50 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2273 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2273 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2273 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2273 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6710 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6710 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1146 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1146 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1146 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1146 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6710 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6710 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4417 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4417 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9980 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9980 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9980 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9980 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4417 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4417 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4417 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4417 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6984 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6984 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2654 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2654 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8324 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8324 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8324 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8324 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2654 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2654 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6984 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6984 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6710 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6710 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4392 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4392 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8917 0.1458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8917 0.1458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4419 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4419 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2654 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2654 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.5096 -0.9090 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 2.5096 -0.9090 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 2.2088 -1.4300 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 2.2088 -1.4300 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 2.7873 -1.2647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7873 -1.2647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.3692 -1.4300 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.3692 -1.4300 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.6701 -0.9090 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | 3.6701 -0.9090 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | 3.0916 -1.0742 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | 3.0916 -1.0742 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | 1.9509 -1.0587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9509 -1.0587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1332 -0.5997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1332 -0.5997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1042 -1.1596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1042 -1.1596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9228 0.0479 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 | + | -3.9228 0.0479 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0 |
| − | -3.5517 -0.4420 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.5517 -0.4420 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -3.0172 -0.2342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -3.0172 -0.2342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.5014 -0.2286 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 | + | -2.5014 -0.2286 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0 |
| − | -2.8762 0.1463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8762 0.1463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3438 -0.1005 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 | + | -3.3438 -0.1005 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0 |
| − | -4.5137 0.0997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5137 0.0997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9020 -0.9032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9020 -0.9032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7109 -0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7109 -0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.3929 0.6564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.3929 0.6564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0437 1.5561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0437 1.5561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5688 -1.7224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5688 -1.7224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5688 -2.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5688 -2.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0362 0.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0362 0.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3184 1.2169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3184 1.2169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 4 3 1 0 0 0 0 | + | 4 3 1 0 0 0 0 |
| − | 1 20 1 0 0 0 0 | + | 1 20 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 21 1 0 0 0 0 | + | 24 21 1 0 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 33 34 1 1 0 0 0 | + | 33 34 1 1 0 0 0 |
| − | 35 34 1 1 0 0 0 | + | 35 34 1 1 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 32 1 0 0 0 0 | + | 37 32 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 40 1 0 0 0 0 | + | 34 40 1 0 0 0 0 |
| − | 35 20 1 0 0 0 0 | + | 35 20 1 0 0 0 0 |
| − | 6 41 1 0 0 0 0 | + | 6 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | 26 43 1 0 0 0 0 | + | 26 43 1 0 0 0 0 |
| − | 43 44 1 0 0 0 0 | + | 43 44 1 0 0 0 0 |
| − | 37 45 1 0 0 0 0 | + | 37 45 1 0 0 0 0 |
| − | 45 46 1 0 0 0 0 | + | 45 46 1 0 0 0 0 |
| − | M STY 1 3 SUP | + | M STY 1 3 SUP |
| − | M SLB 1 3 3 | + | M SLB 1 3 3 |
| − | M SAL 3 2 45 46 | + | M SAL 3 2 45 46 |
| − | M SBL 3 1 49 | + | M SBL 3 1 49 |
| − | M SMT 3 CH2OH | + | M SMT 3 CH2OH |
| − | M SVB 3 49 -3.7422 0.6836 | + | M SVB 3 49 -3.7422 0.6836 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 43 44 | + | M SAL 2 2 43 44 |
| − | M SBL 2 1 47 | + | M SBL 2 1 47 |
| − | M SMT 2 CH2OH | + | M SMT 2 CH2OH |
| − | M SVB 2 47 3.7222 -1.4033 | + | M SVB 2 47 3.7222 -1.4033 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 OCH3 | + | M SMT 1 OCH3 |
| − | M SVB 1 45 -0.3929 0.6564 | + | M SVB 1 45 -0.3929 0.6564 |
| − | S SKP 8 | + | S SKP 8 |
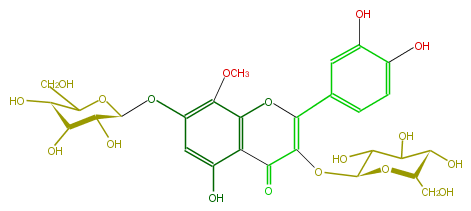
| − | ID FL5FFCGL0007 | + | ID FL5FFCGL0007 |
| − | KNApSAcK_ID C00005707 | + | KNApSAcK_ID C00005707 |
| − | NAME Corniculatusin 3,7-diglucoside | + | NAME Corniculatusin 3,7-diglucoside |
| − | CAS_RN 111133-92-7 | + | CAS_RN 111133-92-7 |
| − | FORMULA C28H32O18 | + | FORMULA C28H32O18 |
| − | EXACTMASS 656.1588642199999 | + | EXACTMASS 656.1588642199999 |
| − | AVERAGEMASS 656.54288 | + | AVERAGEMASS 656.54288 |
| − | SMILES c(O)(c4)c(c2c(c(O[C@@H]([C@@H](O)5)OC(CO)[C@@H]([C@@H]5O)O)4)OC)C(C(O[C@@H](C(O)3)O[C@@H]([C@H](O)C3O)CO)=C(O2)c(c1)cc(c(O)c1)O)=O | + | SMILES c(O)(c4)c(c2c(c(O[C@@H]([C@@H](O)5)OC(CO)[C@@H]([C@@H]5O)O)4)OC)C(C(O[C@@H](C(O)3)O[C@@H]([C@H](O)C3O)CO)=C(O2)c(c1)cc(c(O)c1)O)=O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
46 50 0 0 0 0 0 0 0 0999 V2000
-1.2273 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2273 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6710 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1146 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1146 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6710 0.0833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4417 -1.2014 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9980 -0.8802 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9980 -0.2378 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.4417 0.0833 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4417 -1.7022 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6984 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2654 -0.1204 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8324 0.2070 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8324 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2654 1.1890 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6984 0.8617 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6710 -1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4392 1.2120 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8917 0.1458 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4419 -1.3242 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.2654 1.8435 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.5096 -0.9090 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
2.2088 -1.4300 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
2.7873 -1.2647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.3692 -1.4300 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.6701 -0.9090 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
3.0916 -1.0742 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
1.9509 -1.0587 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1332 -0.5997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1042 -1.1596 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9228 0.0479 0.0000 C 0 0 2 0 0 0 0 0 0 0 0 0
-3.5517 -0.4420 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-3.0172 -0.2342 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.5014 -0.2286 0.0000 C 0 0 1 0 0 0 0 0 0 0 0 0
-2.8762 0.1463 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.3438 -0.1005 0.0000 C 0 0 3 0 0 0 0 0 0 0 0 0
-4.5137 0.0997 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.9020 -0.9032 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7109 -0.7483 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.3929 0.6564 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0437 1.5561 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5688 -1.7224 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5688 -2.5474 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0362 0.4418 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3184 1.2169 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
4 3 1 0 0 0 0
1 20 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 21 1 0 0 0 0
32 33 1 1 0 0 0
33 34 1 1 0 0 0
35 34 1 1 0 0 0
35 36 1 0 0 0 0
36 37 1 0 0 0 0
37 32 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 40 1 0 0 0 0
35 20 1 0 0 0 0
6 41 1 0 0 0 0
41 42 1 0 0 0 0
26 43 1 0 0 0 0
43 44 1 0 0 0 0
37 45 1 0 0 0 0
45 46 1 0 0 0 0
M STY 1 3 SUP
M SLB 1 3 3
M SAL 3 2 45 46
M SBL 3 1 49
M SMT 3 CH2OH
M SVB 3 49 -3.7422 0.6836
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 43 44
M SBL 2 1 47
M SMT 2 CH2OH
M SVB 2 47 3.7222 -1.4033
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 45
M SMT 1 OCH3
M SVB 1 45 -0.3929 0.6564
S SKP 8
ID FL5FFCGL0007
KNApSAcK_ID C00005707
NAME Corniculatusin 3,7-diglucoside
CAS_RN 111133-92-7
FORMULA C28H32O18
EXACTMASS 656.1588642199999
AVERAGEMASS 656.54288
SMILES c(O)(c4)c(c2c(c(O[C@@H]([C@@H](O)5)OC(CO)[C@@H]([C@@H]5O)O)4)OC)C(C(O[C@@H](C(O)3)O[C@@H]([C@H](O)C3O)CO)=C(O2)c(c1)cc(c(O)c1)O)=O
M END