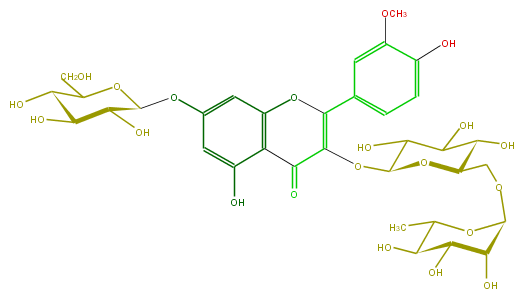
Mol:FL5FADGL0027
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 55 60 0 0 0 0 0 0 0 0999 V2000 | + | 55 60 0 0 0 0 0 0 0 0999 V2000 |
| − | -1.2046 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2046 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2046 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2046 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4933 -0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4933 -0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2179 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2179 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2179 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2179 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4933 0.6487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4933 0.6487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9292 -0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9292 -0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6405 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6405 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6405 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6405 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9292 0.6487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9292 0.6487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9292 -1.6343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9292 -1.6343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3515 0.6486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3515 0.6486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0764 0.2300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0764 0.2300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8012 0.6486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8012 0.6486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8012 1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8012 1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0764 1.9042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0764 1.9042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3515 1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3515 1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9156 0.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9156 0.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4346 -0.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4346 -0.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4933 -1.8149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4933 -1.8149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.2262 -2.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.2262 -2.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7969 -3.0417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7969 -3.0417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.6224 -2.8058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.6224 -2.8058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4528 -3.0417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4528 -3.0417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8821 -2.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8821 -2.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0566 -2.5341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0566 -2.5341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4936 -2.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4936 -2.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5871 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5871 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1579 -1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1579 -1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9833 -0.8879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9833 -0.8879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8137 -1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8137 -1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.2431 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.2431 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4175 -0.6162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4175 -0.6162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6676 -0.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6676 -0.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.8835 -0.0161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.8835 -0.0161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.8630 -0.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.8630 -0.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4384 -0.9616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4384 -0.9616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.7481 -1.4556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.7481 -1.4556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1382 -2.8864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1382 -2.8864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.1674 -3.4697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.1674 -3.4697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.4528 -3.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.4528 -3.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4767 1.9172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4767 1.9172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7799 0.7644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7799 0.7644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2340 0.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2340 0.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4479 0.3495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4479 0.3495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6895 0.3577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6895 0.3577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2407 0.9090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2407 0.9090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.0436 0.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.0436 0.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.5216 0.4944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.5216 0.4944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0366 0.1272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0366 0.1272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7271 -0.1604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7271 -0.1604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.6317 1.1555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.6317 1.1555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.8821 1.4223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.8821 1.4223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0689 2.6187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0689 2.6187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6332 3.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6332 3.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 1 18 1 0 0 0 0 | + | 1 18 1 0 0 0 0 |
| − | 8 19 1 0 0 0 0 | + | 8 19 1 0 0 0 0 |
| − | 3 20 1 0 0 0 0 | + | 3 20 1 0 0 0 0 |
| − | 21 22 1 0 0 0 0 | + | 21 22 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 21 1 0 0 0 0 | + | 26 21 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 1 0 0 0 | + | 29 30 1 1 0 0 0 |
| − | 30 31 1 1 0 0 0 | + | 30 31 1 1 0 0 0 |
| − | 32 31 1 1 0 0 0 | + | 32 31 1 1 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 28 1 0 0 0 0 | + | 33 28 1 0 0 0 0 |
| − | 28 34 1 0 0 0 0 | + | 28 34 1 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 1 0 0 0 0 | + | 32 36 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 25 38 1 0 0 0 0 | + | 25 38 1 0 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 22 39 1 0 0 0 0 | + | 22 39 1 0 0 0 0 |
| − | 23 40 1 0 0 0 0 | + | 23 40 1 0 0 0 0 |
| − | 24 41 1 0 0 0 0 | + | 24 41 1 0 0 0 0 |
| − | 29 19 1 0 0 0 0 | + | 29 19 1 0 0 0 0 |
| − | 42 15 1 0 0 0 0 | + | 42 15 1 0 0 0 0 |
| − | 43 44 1 1 0 0 0 | + | 43 44 1 1 0 0 0 |
| − | 44 45 1 1 0 0 0 | + | 44 45 1 1 0 0 0 |
| − | 46 45 1 1 0 0 0 | + | 46 45 1 1 0 0 0 |
| − | 46 47 1 0 0 0 0 | + | 46 47 1 0 0 0 0 |
| − | 47 48 1 0 0 0 0 | + | 47 48 1 0 0 0 0 |
| − | 48 43 1 0 0 0 0 | + | 48 43 1 0 0 0 0 |
| − | 43 49 1 0 0 0 0 | + | 43 49 1 0 0 0 0 |
| − | 44 50 1 0 0 0 0 | + | 44 50 1 0 0 0 0 |
| − | 45 51 1 0 0 0 0 | + | 45 51 1 0 0 0 0 |
| − | 46 18 1 0 0 0 0 | + | 46 18 1 0 0 0 0 |
| − | 52 53 1 0 0 0 0 | + | 52 53 1 0 0 0 0 |
| − | 48 52 1 0 0 0 0 | + | 48 52 1 0 0 0 0 |
| − | 54 55 1 0 0 0 0 | + | 54 55 1 0 0 0 0 |
| − | 16 54 1 0 0 0 0 | + | 16 54 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 52 53 | + | M SAL 1 2 52 53 |
| − | M SBL 1 1 58 | + | M SBL 1 1 58 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 58 0.5881 -0.5347 | + | M SBV 1 58 0.5881 -0.5347 |
| − | M STY 1 2 SUP | + | M STY 1 2 SUP |
| − | M SLB 1 2 2 | + | M SLB 1 2 2 |
| − | M SAL 2 2 54 55 | + | M SAL 2 2 54 55 |
| − | M SBL 2 1 60 | + | M SBL 2 1 60 |
| − | M SMT 2 OCH3 | + | M SMT 2 OCH3 |
| − | M SBV 2 60 0.0075 -0.7146 | + | M SBV 2 60 0.0075 -0.7146 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL5FADGL0027 | + | ID FL5FADGL0027 |
| − | FORMULA C34H42O21 | + | FORMULA C34H42O21 |
| − | EXACTMASS 786.2218584059999 | + | EXACTMASS 786.2218584059999 |
| − | AVERAGEMASS 786.68468 | + | AVERAGEMASS 786.68468 |
| − | SMILES C(=C(c(c6)cc(c(c6)O)OC)3)(OC(O4)C(C(O)C(C4COC(C5O)OC(C)C(C5O)O)O)O)C(=O)c(c1O)c(O3)cc(OC(C2O)OC(C(C(O)2)O)CO)c1 | + | SMILES C(=C(c(c6)cc(c(c6)O)OC)3)(OC(O4)C(C(O)C(C4COC(C5O)OC(C)C(C5O)O)O)O)C(=O)c(c1O)c(O3)cc(OC(C2O)OC(C(C(O)2)O)CO)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
55 60 0 0 0 0 0 0 0 0999 V2000
-1.2046 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2046 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4933 -0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2179 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2179 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4933 0.6487 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9292 -0.9938 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6405 -0.5833 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6405 0.2381 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9292 0.6487 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9292 -1.6343 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3515 0.6486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0764 0.2300 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8012 0.6486 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8012 1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0764 1.9042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.3515 1.4856 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9156 0.6486 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4346 -0.9788 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4933 -1.8149 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.2262 -2.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.7969 -3.0417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.6224 -2.8058 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.4528 -3.0417 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.8821 -2.2983 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.0566 -2.5341 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.4936 -2.4093 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5871 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1579 -1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.9833 -0.8879 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8137 -1.1238 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.2431 -0.3805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4175 -0.6162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6676 -0.5129 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.8835 -0.0161 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.8630 -0.4707 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4384 -0.9616 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.7481 -1.4556 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1382 -2.8864 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.1674 -3.4697 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.4528 -3.7660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.4767 1.9172 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7799 0.7644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2340 0.0438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.4479 0.3495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6895 0.3577 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2407 0.9090 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.0436 0.6207 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.5216 0.4944 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.0366 0.1272 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7271 -0.1604 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.6317 1.1555 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.8821 1.4223 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0689 2.6187 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6332 3.7660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
1 18 1 0 0 0 0
8 19 1 0 0 0 0
3 20 1 0 0 0 0
21 22 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 21 1 0 0 0 0
21 27 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 1 0 0 0
30 31 1 1 0 0 0
32 31 1 1 0 0 0
32 33 1 0 0 0 0
33 28 1 0 0 0 0
28 34 1 0 0 0 0
33 35 1 0 0 0 0
32 36 1 0 0 0 0
31 37 1 0 0 0 0
25 38 1 0 0 0 0
37 38 1 0 0 0 0
22 39 1 0 0 0 0
23 40 1 0 0 0 0
24 41 1 0 0 0 0
29 19 1 0 0 0 0
42 15 1 0 0 0 0
43 44 1 1 0 0 0
44 45 1 1 0 0 0
46 45 1 1 0 0 0
46 47 1 0 0 0 0
47 48 1 0 0 0 0
48 43 1 0 0 0 0
43 49 1 0 0 0 0
44 50 1 0 0 0 0
45 51 1 0 0 0 0
46 18 1 0 0 0 0
52 53 1 0 0 0 0
48 52 1 0 0 0 0
54 55 1 0 0 0 0
16 54 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 52 53
M SBL 1 1 58
M SMT 1 ^ CH2OH
M SBV 1 58 0.5881 -0.5347
M STY 1 2 SUP
M SLB 1 2 2
M SAL 2 2 54 55
M SBL 2 1 60
M SMT 2 OCH3
M SBV 2 60 0.0075 -0.7146
S SKP 5
ID FL5FADGL0027
FORMULA C34H42O21
EXACTMASS 786.2218584059999
AVERAGEMASS 786.68468
SMILES C(=C(c(c6)cc(c(c6)O)OC)3)(OC(O4)C(C(O)C(C4COC(C5O)OC(C)C(C5O)O)O)O)C(=O)c(c1O)c(O3)cc(OC(C2O)OC(C(C(O)2)O)CO)c1
M END