Mol:FL5FACGS0112
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 68 74 0 0 0 0 0 0 0 0999 V2000 | + | 68 74 0 0 0 0 0 0 0 0999 V2000 |
| − | 3.8757 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8757 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1613 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1613 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1613 3.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1613 3.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8757 4.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8757 4.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5902 3.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5902 3.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.5902 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.5902 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4468 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4468 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7323 3.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7323 3.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0179 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0179 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0179 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0179 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7323 1.4713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7323 1.4713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4468 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4468 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3034 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3034 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4111 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4111 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4111 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4111 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3034 1.4713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3034 1.4713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7323 0.8105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7323 0.8105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1256 3.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1256 3.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0458 1.4383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0458 1.4383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3034 0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3034 0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1821 4.2880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1821 4.2880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.8757 5.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.8757 5.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4567 -0.4994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4567 -0.4994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2508 -0.7239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2508 -0.7239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6772 -0.0173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6772 -0.0173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.4041 0.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.4041 0.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6100 0.5980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6100 0.5980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1835 -0.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1835 -0.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6625 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6625 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.1254 0.0659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.1254 0.0659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4472 -1.1827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4472 -1.1827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7485 -0.8511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7485 -0.8511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.0005 -0.5956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.0005 -0.5956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9597 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9597 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7705 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7705 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.1334 -1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.1334 -1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8234 -1.4805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8234 -1.4805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0126 -1.3262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0126 -1.3262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6496 -2.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6496 -2.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0639 -1.7846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0639 -1.7846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5406 -1.9672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5406 -1.9672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4694 -2.5565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4694 -2.5565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0298 -3.1784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0298 -3.1784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7239 -2.2183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7239 -2.2183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9414 -4.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9414 -4.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7640 -4.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7640 -4.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0454 -3.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0454 -3.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6828 -3.1788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6828 -3.1788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8602 -3.1123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8602 -3.1123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5787 -3.8882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5787 -3.8882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0074 -3.6846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0074 -3.6846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5069 -3.9221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5069 -3.9221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0646 -5.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0646 -5.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.2960 -4.7009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.2960 -4.7009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4746 -4.2076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4746 -4.2076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.5805 -3.4214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.5805 -3.4214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1686 -2.7079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1686 -2.7079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4044 -3.4214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4044 -3.4214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8164 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8164 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6403 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6403 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0528 -3.4205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0528 -3.4205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8778 -3.4205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8778 -3.4205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.2903 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.2903 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.8778 -4.8494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.8778 -4.8494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0528 -4.8494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0528 -4.8494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1141 -4.1350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1141 -4.1350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.4559 -2.8423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.4559 -2.8423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1821 -2.8423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1821 -2.8423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 2 0 0 0 0 | + | 1 2 2 0 0 0 0 |
| − | 2 3 1 0 0 0 0 | + | 2 3 1 0 0 0 0 |
| − | 3 4 2 0 0 0 0 | + | 3 4 2 0 0 0 0 |
| − | 4 5 1 0 0 0 0 | + | 4 5 1 0 0 0 0 |
| − | 5 6 2 0 0 0 0 | + | 5 6 2 0 0 0 0 |
| − | 6 1 1 0 0 0 0 | + | 6 1 1 0 0 0 0 |
| − | 2 7 1 0 0 0 0 | + | 2 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 1 0 0 0 0 | + | 8 9 1 0 0 0 0 |
| − | 9 10 2 0 0 0 0 | + | 9 10 2 0 0 0 0 |
| − | 10 11 1 0 0 0 0 | + | 10 11 1 0 0 0 0 |
| − | 11 12 1 0 0 0 0 | + | 11 12 1 0 0 0 0 |
| − | 12 7 2 0 0 0 0 | + | 12 7 2 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 10 1 0 0 0 0 | + | 16 10 1 0 0 0 0 |
| − | 14 18 1 0 0 0 0 | + | 14 18 1 0 0 0 0 |
| − | 11 17 2 0 0 0 0 | + | 11 17 2 0 0 0 0 |
| − | 12 19 1 0 0 0 0 | + | 12 19 1 0 0 0 0 |
| − | 16 20 1 0 0 0 0 | + | 16 20 1 0 0 0 0 |
| − | 5 21 1 0 0 0 0 | + | 5 21 1 0 0 0 0 |
| − | 4 22 1 0 0 0 0 | + | 4 22 1 0 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 26 25 1 1 0 0 0 | + | 26 25 1 1 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 28 29 1 0 0 0 0 | + | 28 29 1 0 0 0 0 |
| − | 29 30 1 0 0 0 0 | + | 29 30 1 0 0 0 0 |
| − | 23 31 1 0 0 0 0 | + | 23 31 1 0 0 0 0 |
| − | 24 32 1 0 0 0 0 | + | 24 32 1 0 0 0 0 |
| − | 25 33 1 0 0 0 0 | + | 25 33 1 0 0 0 0 |
| − | 26 19 1 0 0 0 0 | + | 26 19 1 0 0 0 0 |
| − | 34 35 1 1 0 0 0 | + | 34 35 1 1 0 0 0 |
| − | 35 36 1 1 0 0 0 | + | 35 36 1 1 0 0 0 |
| − | 37 36 1 1 0 0 0 | + | 37 36 1 1 0 0 0 |
| − | 37 38 1 0 0 0 0 | + | 37 38 1 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 34 1 0 0 0 0 | + | 39 34 1 0 0 0 0 |
| − | 39 40 1 0 0 0 0 | + | 39 40 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 34 42 1 0 0 0 0 | + | 34 42 1 0 0 0 0 |
| − | 35 43 1 0 0 0 0 | + | 35 43 1 0 0 0 0 |
| − | 36 44 1 0 0 0 0 | + | 36 44 1 0 0 0 0 |
| − | 37 33 1 0 0 0 0 | + | 37 33 1 0 0 0 0 |
| − | 45 46 1 1 0 0 0 | + | 45 46 1 1 0 0 0 |
| − | 46 47 1 1 0 0 0 | + | 46 47 1 1 0 0 0 |
| − | 48 47 1 1 0 0 0 | + | 48 47 1 1 0 0 0 |
| − | 48 49 1 0 0 0 0 | + | 48 49 1 0 0 0 0 |
| − | 49 50 1 0 0 0 0 | + | 49 50 1 0 0 0 0 |
| − | 50 45 1 0 0 0 0 | + | 50 45 1 0 0 0 0 |
| − | 50 51 1 0 0 0 0 | + | 50 51 1 0 0 0 0 |
| − | 51 52 1 0 0 0 0 | + | 51 52 1 0 0 0 0 |
| − | 45 53 1 0 0 0 0 | + | 45 53 1 0 0 0 0 |
| − | 46 54 1 0 0 0 0 | + | 46 54 1 0 0 0 0 |
| − | 47 55 1 0 0 0 0 | + | 47 55 1 0 0 0 0 |
| − | 48 44 1 0 0 0 0 | + | 48 44 1 0 0 0 0 |
| − | 56 57 2 0 0 0 0 | + | 56 57 2 0 0 0 0 |
| − | 56 58 1 0 0 0 0 | + | 56 58 1 0 0 0 0 |
| − | 58 59 2 0 0 0 0 | + | 58 59 2 0 0 0 0 |
| − | 59 60 1 0 0 0 0 | + | 59 60 1 0 0 0 0 |
| − | 60 61 2 0 0 0 0 | + | 60 61 2 0 0 0 0 |
| − | 61 62 1 0 0 0 0 | + | 61 62 1 0 0 0 0 |
| − | 62 63 2 0 0 0 0 | + | 62 63 2 0 0 0 0 |
| − | 63 64 1 0 0 0 0 | + | 63 64 1 0 0 0 0 |
| − | 64 65 2 0 0 0 0 | + | 64 65 2 0 0 0 0 |
| − | 65 60 1 0 0 0 0 | + | 65 60 1 0 0 0 0 |
| − | 63 66 1 0 0 0 0 | + | 63 66 1 0 0 0 0 |
| − | 62 67 1 0 0 0 0 | + | 62 67 1 0 0 0 0 |
| − | 67 68 1 0 0 0 0 | + | 67 68 1 0 0 0 0 |
| − | 52 56 1 0 0 0 0 | + | 52 56 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGS0112 | + | ID FL5FACGS0112 |
| − | KNApSAcK_ID C00013884 | + | KNApSAcK_ID C00013884 |
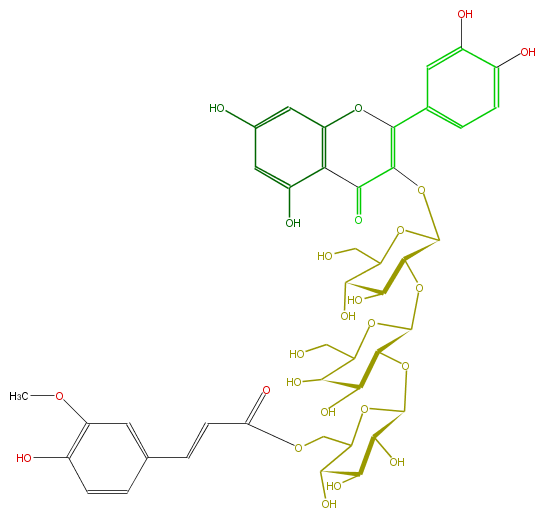
| − | NAME Quercetin 3-(6''''-feruloylglucosyl)-(1->2)-galactosyl-(1->2)-glucoside;3-[[O-6-O-[(2E)-3-(4-Hydroxy-3-methoxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl-(1->2)-O-beta-D-galactopyranosyl-(1->2)-beta-D-glucopyranosyl]oxy]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-1-benzopyran-4-one | + | NAME Quercetin 3-(6''''-feruloylglucosyl)-(1->2)-galactosyl-(1->2)-glucoside;3-[[O-6-O-[(2E)-3-(4-Hydroxy-3-methoxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl-(1->2)-O-beta-D-galactopyranosyl-(1->2)-beta-D-glucopyranosyl]oxy]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-1-benzopyran-4-one |
| − | CAS_RN 197294-29-4 | + | CAS_RN 197294-29-4 |
| − | FORMULA C43H48O25 | + | FORMULA C43H48O25 |
| − | EXACTMASS 964.2484670859999 | + | EXACTMASS 964.2484670859999 |
| − | AVERAGEMASS 964.82622 | + | AVERAGEMASS 964.82622 |
| − | SMILES c(c1C=CC(=O)OCC(C2O)OC(OC(C3OC(C4O)C(OC(C(=O)5)=C(c(c7)cc(c(c7)O)O)Oc(c6)c5c(O)cc(O)6)OC(C4O)CO)C(O)C(O)C(O3)CO)C(O)C2O)c(OC)c(cc1)O | + | SMILES c(c1C=CC(=O)OCC(C2O)OC(OC(C3OC(C4O)C(OC(C(=O)5)=C(c(c7)cc(c(c7)O)O)Oc(c6)c5c(O)cc(O)6)OC(C4O)CO)C(O)C(O)C(O3)CO)C(O)C2O)c(OC)c(cc1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
68 74 0 0 0 0 0 0 0 0999 V2000
3.8757 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1613 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1613 3.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.8757 4.3588 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5902 3.9463 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5902 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4468 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7323 3.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0179 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0179 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7323 1.4713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4468 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3034 3.1213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4111 2.7088 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4111 1.8838 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3034 1.4713 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7323 0.8105 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.1256 3.1213 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0458 1.4383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3034 0.7446 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.1821 4.2880 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.8757 5.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4567 -0.4994 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2508 -0.7239 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6772 -0.0173 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4041 0.3734 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6100 0.5980 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.1835 -0.1086 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6625 0.2019 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.1254 0.0659 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4472 -1.1827 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7485 -0.8511 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.0005 -0.5956 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9597 -2.5203 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7705 -2.6745 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.1334 -1.9333 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8234 -1.4805 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0126 -1.3262 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.6496 -2.0674 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0639 -1.7846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5406 -1.9672 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4694 -2.5565 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0298 -3.1784 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7239 -2.2183 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9414 -4.4124 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7640 -4.4788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0454 -3.7030 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6828 -3.1788 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.8602 -3.1123 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.5787 -3.8882 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0074 -3.6846 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5069 -3.9221 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0646 -5.0846 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.2960 -4.7009 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4746 -4.2076 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.5805 -3.4214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1686 -2.7079 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4044 -3.4214 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8164 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.6403 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0528 -3.4205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8778 -3.4205 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.2903 -4.1350 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.8778 -4.8494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0528 -4.8494 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1141 -4.1350 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.4559 -2.8423 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-5.1821 -2.8423 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 2 0 0 0 0
2 3 1 0 0 0 0
3 4 2 0 0 0 0
4 5 1 0 0 0 0
5 6 2 0 0 0 0
6 1 1 0 0 0 0
2 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 1 0 0 0 0
9 10 2 0 0 0 0
10 11 1 0 0 0 0
11 12 1 0 0 0 0
12 7 2 0 0 0 0
9 13 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 10 1 0 0 0 0
14 18 1 0 0 0 0
11 17 2 0 0 0 0
12 19 1 0 0 0 0
16 20 1 0 0 0 0
5 21 1 0 0 0 0
4 22 1 0 0 0 0
23 24 1 1 0 0 0
24 25 1 1 0 0 0
26 25 1 1 0 0 0
26 27 1 0 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
28 29 1 0 0 0 0
29 30 1 0 0 0 0
23 31 1 0 0 0 0
24 32 1 0 0 0 0
25 33 1 0 0 0 0
26 19 1 0 0 0 0
34 35 1 1 0 0 0
35 36 1 1 0 0 0
37 36 1 1 0 0 0
37 38 1 0 0 0 0
38 39 1 0 0 0 0
39 34 1 0 0 0 0
39 40 1 0 0 0 0
40 41 1 0 0 0 0
34 42 1 0 0 0 0
35 43 1 0 0 0 0
36 44 1 0 0 0 0
37 33 1 0 0 0 0
45 46 1 1 0 0 0
46 47 1 1 0 0 0
48 47 1 1 0 0 0
48 49 1 0 0 0 0
49 50 1 0 0 0 0
50 45 1 0 0 0 0
50 51 1 0 0 0 0
51 52 1 0 0 0 0
45 53 1 0 0 0 0
46 54 1 0 0 0 0
47 55 1 0 0 0 0
48 44 1 0 0 0 0
56 57 2 0 0 0 0
56 58 1 0 0 0 0
58 59 2 0 0 0 0
59 60 1 0 0 0 0
60 61 2 0 0 0 0
61 62 1 0 0 0 0
62 63 2 0 0 0 0
63 64 1 0 0 0 0
64 65 2 0 0 0 0
65 60 1 0 0 0 0
63 66 1 0 0 0 0
62 67 1 0 0 0 0
67 68 1 0 0 0 0
52 56 1 0 0 0 0
S SKP 8
ID FL5FACGS0112
KNApSAcK_ID C00013884
NAME Quercetin 3-(6''''-feruloylglucosyl)-(1->2)-galactosyl-(1->2)-glucoside;3-[[O-6-O-[(2E)-3-(4-Hydroxy-3-methoxyphenyl)-1-oxo-2-propenyl]-beta-D-glucopyranosyl-(1->2)-O-beta-D-galactopyranosyl-(1->2)-beta-D-glucopyranosyl]oxy]-2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4H-1-benzopyran-4-one
CAS_RN 197294-29-4
FORMULA C43H48O25
EXACTMASS 964.2484670859999
AVERAGEMASS 964.82622
SMILES c(c1C=CC(=O)OCC(C2O)OC(OC(C3OC(C4O)C(OC(C(=O)5)=C(c(c7)cc(c(c7)O)O)Oc(c6)c5c(O)cc(O)6)OC(C4O)CO)C(O)C(O)C(O3)CO)C(O)C2O)c(OC)c(cc1)O
M END