Mol:FL5FACGA0035
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 42 45 0 0 0 0 0 0 0 0999 V2000 | + | 42 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.3850 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3850 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3850 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3850 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8287 0.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8287 0.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2724 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2724 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2724 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2724 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8287 1.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8287 1.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7161 0.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7161 0.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1598 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1598 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1598 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1598 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7161 1.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7161 1.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7161 -0.2693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7161 -0.2693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3963 1.5162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3963 1.5162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9633 1.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9633 1.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5302 1.5162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5302 1.5162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5302 2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5302 2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9633 2.4982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9633 2.4982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3963 2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3963 2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8287 -0.4106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8287 -0.4106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3239 2.5502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3239 2.5502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8800 1.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8800 1.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.4540 0.1984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.4540 0.1984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9633 3.1527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9633 3.1527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0436 -0.8839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0436 -0.8839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3721 -0.4963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3721 -0.4963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.5852 -1.2418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.5852 -1.2418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.3721 -1.9917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.3721 -1.9917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0436 -2.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0436 -2.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8306 -1.6339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8306 -1.6339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.8507 -0.1640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.8507 -0.1640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3622 -1.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3622 -1.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.8138 -2.1731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.8138 -2.1731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3022 -2.6614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3022 -2.6614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9454 -1.6042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9454 -1.6042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.9454 -1.2163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.9454 -1.2163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4626 -1.9028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4626 -1.9028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0185 -3.1527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0185 -3.1527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8800 -2.6614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8800 -2.6614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.5179 0.2213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.5179 0.2213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3911 0.6945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3911 0.6945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0037 -0.0592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0037 -0.0592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.0492 -2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.0492 -2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7636 -2.6303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7636 -2.6303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 3 18 1 0 0 0 0 | + | 3 18 1 0 0 0 0 |
| − | 19 15 1 0 0 0 0 | + | 19 15 1 0 0 0 0 |
| − | 20 1 1 0 0 0 0 | + | 20 1 1 0 0 0 0 |
| − | 21 8 1 0 0 0 0 | + | 21 8 1 0 0 0 0 |
| − | 16 22 1 0 0 0 0 | + | 16 22 1 0 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 1 0 0 0 | + | 24 25 1 1 0 0 0 |
| − | 25 26 1 1 0 0 0 | + | 25 26 1 1 0 0 0 |
| − | 27 26 1 1 0 0 0 | + | 27 26 1 1 0 0 0 |
| − | 27 28 1 0 0 0 0 | + | 27 28 1 0 0 0 0 |
| − | 28 23 1 0 0 0 0 | + | 28 23 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 28 30 1 0 0 0 0 | + | 28 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 24 21 1 0 0 0 0 | + | 24 21 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 30 33 1 0 0 0 0 | + | 30 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 32 36 2 0 0 0 0 | + | 32 36 2 0 0 0 0 |
| − | 32 37 1 0 0 0 0 | + | 32 37 1 0 0 0 0 |
| − | 29 38 1 0 0 0 0 | + | 29 38 1 0 0 0 0 |
| − | 38 39 2 0 0 0 0 | + | 38 39 2 0 0 0 0 |
| − | 38 40 1 0 0 0 0 | + | 38 40 1 0 0 0 0 |
| − | 26 41 1 0 0 0 0 | + | 26 41 1 0 0 0 0 |
| − | 41 42 1 0 0 0 0 | + | 41 42 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 41 42 | + | M SAL 1 2 41 42 |
| − | M SBL 1 1 44 | + | M SBL 1 1 44 |
| − | M SMT 1 CH2OH | + | M SMT 1 CH2OH |
| − | M SBV 1 44 -5.1376 3.6503 | + | M SBV 1 44 -5.1376 3.6503 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL5FACGA0035 | + | ID FL5FACGA0035 |
| − | KNApSAcK_ID C00005954 | + | KNApSAcK_ID C00005954 |
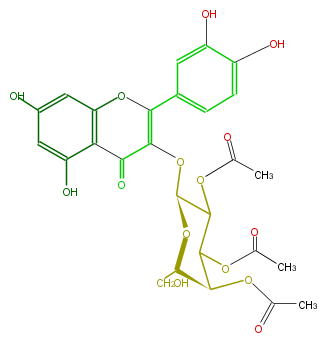
| − | NAME Quercetin 3-(2'',3'',4''-triacetylgalactoside) | + | NAME Quercetin 3-(2'',3'',4''-triacetylgalactoside) |
| − | CAS_RN 146018-89-5 | + | CAS_RN 146018-89-5 |
| − | FORMULA C27H26O15 | + | FORMULA C27H26O15 |
| − | EXACTMASS 590.127170162 | + | EXACTMASS 590.127170162 |
| − | AVERAGEMASS 590.4863399999999 | + | AVERAGEMASS 590.4863399999999 |
| − | SMILES c(c1O)c(c(C(=O)2)c(OC(c(c4)ccc(O)c4O)=C2OC(O3)C(C(C(C(CO)3)OC(C)=O)OC(C)=O)OC(C)=O)c1)O | + | SMILES c(c1O)c(c(C(=O)2)c(OC(c(c4)ccc(O)c4O)=C2OC(O3)C(C(C(C(CO)3)OC(C)=O)OC(C)=O)OC(C)=O)c1)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
42 45 0 0 0 0 0 0 0 0999 V2000
-2.3850 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3850 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8287 0.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2724 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2724 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8287 1.5163 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7161 0.2316 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1598 0.5527 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1598 1.1951 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7161 1.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.7161 -0.2693 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3963 1.5162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9633 1.1888 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5302 1.5162 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5302 2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9633 2.4982 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3963 2.1709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8287 -0.4106 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3239 2.5502 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.8800 1.5163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.4540 0.1984 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9633 3.1527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0436 -0.8839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.3721 -0.4963 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.5852 -1.2418 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.3721 -1.9917 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0436 -2.3795 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8306 -1.6339 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8507 -0.1640 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3622 -1.9409 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.8138 -2.1731 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3022 -2.6614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9454 -1.6042 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.9454 -1.2163 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.4626 -1.9028 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0185 -3.1527 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.8800 -2.6614 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5179 0.2213 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3911 0.6945 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.0037 -0.0592 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0492 -2.2178 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7636 -2.6303 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
3 18 1 0 0 0 0
19 15 1 0 0 0 0
20 1 1 0 0 0 0
21 8 1 0 0 0 0
16 22 1 0 0 0 0
23 24 1 0 0 0 0
24 25 1 1 0 0 0
25 26 1 1 0 0 0
27 26 1 1 0 0 0
27 28 1 0 0 0 0
28 23 1 0 0 0 0
23 29 1 0 0 0 0
28 30 1 0 0 0 0
27 31 1 0 0 0 0
24 21 1 0 0 0 0
31 32 1 0 0 0 0
30 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
32 36 2 0 0 0 0
32 37 1 0 0 0 0
29 38 1 0 0 0 0
38 39 2 0 0 0 0
38 40 1 0 0 0 0
26 41 1 0 0 0 0
41 42 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 41 42
M SBL 1 1 44
M SMT 1 CH2OH
M SBV 1 44 -5.1376 3.6503
S SKP 8
ID FL5FACGA0035
KNApSAcK_ID C00005954
NAME Quercetin 3-(2'',3'',4''-triacetylgalactoside)
CAS_RN 146018-89-5
FORMULA C27H26O15
EXACTMASS 590.127170162
AVERAGEMASS 590.4863399999999
SMILES c(c1O)c(c(C(=O)2)c(OC(c(c4)ccc(O)c4O)=C2OC(O3)C(C(C(C(CO)3)OC(C)=O)OC(C)=O)OC(C)=O)c1)O
M END