Mol:FL5FAAGSS001
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 37 40 0 0 0 0 0 0 0 0999 V2000 | + | 37 40 0 0 0 0 0 0 0 0999 V2000 |
| − | -2.5927 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5927 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.5927 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.5927 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8783 0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8783 0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1639 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1639 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.1639 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.1639 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8783 1.8283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8783 1.8283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4494 0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4494 0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2649 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2649 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.2650 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.2650 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4494 1.8283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4494 1.8283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4493 -0.4647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4493 -0.4647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9792 1.8283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9792 1.8283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7073 1.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7073 1.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4355 1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4355 1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.4355 2.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.4355 2.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7072 3.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7072 3.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.9792 2.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.9792 2.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.0485 0.1250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.0485 0.1250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.3007 -0.3038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.3007 -0.3038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.4586 -0.6101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.4586 -0.6101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2769 -0.9754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2769 -0.9754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.7827 -1.7212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.7827 -1.7212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6249 -1.4148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6249 -1.4148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.8065 -1.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.8065 -1.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.6763 0.0568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.6763 0.0568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6217 -0.8311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6217 -0.8311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.9236 -1.7136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.9236 -1.7136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1633 3.0893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1633 3.0893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.8783 -0.4636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.8783 -0.4636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.7947 1.7939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.7947 1.7939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1286 1.7939 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1286 1.7939 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3116 1.7939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3116 1.7939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1286 2.6051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1286 2.6051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1286 1.0057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1286 1.0057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.7086 -2.1502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.7086 -2.1502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.7947 -1.7549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.7947 -1.7549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5430 -3.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5430 -3.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 19 20 1 0 0 0 0 | + | 19 20 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 19 1 0 0 0 0 | + | 24 19 1 0 0 0 0 |
| − | 19 25 1 0 0 0 0 | + | 19 25 1 0 0 0 0 |
| − | 24 26 1 0 0 0 0 | + | 24 26 1 0 0 0 0 |
| − | 23 27 1 0 0 0 0 | + | 23 27 1 0 0 0 0 |
| − | 20 18 1 0 0 0 0 | + | 20 18 1 0 0 0 0 |
| − | 18 8 1 0 0 0 0 | + | 18 8 1 0 0 0 0 |
| − | 22 21 1 1 0 0 0 | + | 22 21 1 1 0 0 0 |
| − | 15 28 1 0 0 0 0 | + | 15 28 1 0 0 0 0 |
| − | 3 29 1 0 0 0 0 | + | 3 29 1 0 0 0 0 |
| − | 30 31 1 0 0 0 0 | + | 30 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 31 33 2 0 0 0 0 | + | 31 33 2 0 0 0 0 |
| − | 31 34 2 0 0 0 0 | + | 31 34 2 0 0 0 0 |
| − | 1 32 1 0 0 0 0 | + | 1 32 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 35 37 1 0 0 0 0 | + | 35 37 1 0 0 0 0 |
| − | 22 35 1 0 0 0 0 | + | 22 35 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 3 35 36 37 | + | M SAL 1 3 35 36 37 |
| − | M SBL 1 1 40 | + | M SBL 1 1 40 |
| − | M SMT 1 COOH | + | M SMT 1 COOH |
| − | M SBV 1 40 -0.9259 0.4290 | + | M SBV 1 40 -0.9259 0.4290 |
| − | S SKP 5 | + | S SKP 5 |
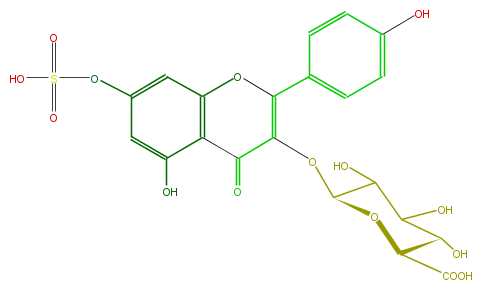
| − | ID FL5FAAGSS001 | + | ID FL5FAAGSS001 |
| − | FORMULA C21H18O15S | + | FORMULA C21H18O15S |
| − | EXACTMASS 542.036640596 | + | EXACTMASS 542.036640596 |
| − | AVERAGEMASS 542.42462 | + | AVERAGEMASS 542.42462 |
| − | SMILES C(C(C(O)=O)1)(O)C(C(C(OC(C3=O)=C(c(c4)ccc(O)c4)Oc(c23)cc(cc2O)OS(O)(=O)=O)O1)O)O | + | SMILES C(C(C(O)=O)1)(O)C(C(C(OC(C3=O)=C(c(c4)ccc(O)c4)Oc(c23)cc(cc2O)OS(O)(=O)=O)O1)O)O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
37 40 0 0 0 0 0 0 0 0999 V2000
-2.5927 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5927 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8783 0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1639 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.1639 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.8783 1.8283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4494 0.1784 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2649 0.5910 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.2650 1.4159 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.4494 1.8283 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4493 -0.4647 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.9792 1.8283 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7073 1.4077 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4355 1.8282 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4355 2.6689 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7072 3.0894 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.9792 2.6690 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0485 0.1250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.3007 -0.3038 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.4586 -0.6101 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2769 -0.9754 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.7827 -1.7212 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6249 -1.4148 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.8065 -1.0495 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.6763 0.0568 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6217 -0.8311 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.9236 -1.7136 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.1633 3.0893 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.8783 -0.4636 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.7947 1.7939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1286 1.7939 0.0000 S 0 0 0 0 0 0 0 0 0 0 0 0
-3.3116 1.7939 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1286 2.6051 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.1286 1.0057 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.7086 -2.1502 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.7947 -1.7549 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5430 -3.0894 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
19 20 1 0 0 0 0
20 21 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 19 1 0 0 0 0
19 25 1 0 0 0 0
24 26 1 0 0 0 0
23 27 1 0 0 0 0
20 18 1 0 0 0 0
18 8 1 0 0 0 0
22 21 1 1 0 0 0
15 28 1 0 0 0 0
3 29 1 0 0 0 0
30 31 1 0 0 0 0
31 32 1 0 0 0 0
31 33 2 0 0 0 0
31 34 2 0 0 0 0
1 32 1 0 0 0 0
35 36 2 0 0 0 0
35 37 1 0 0 0 0
22 35 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 3 35 36 37
M SBL 1 1 40
M SMT 1 COOH
M SBV 1 40 -0.9259 0.4290
S SKP 5
ID FL5FAAGSS001
FORMULA C21H18O15S
EXACTMASS 542.036640596
AVERAGEMASS 542.42462
SMILES C(C(C(O)=O)1)(O)C(C(C(OC(C3=O)=C(c(c4)ccc(O)c4)Oc(c23)cc(cc2O)OS(O)(=O)=O)O1)O)O
M END