Mol:FL3FACGS0062
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 43 47 0 0 0 0 0 0 0 0999 V2000 | + | 43 47 0 0 0 0 0 0 0 0999 V2000 |
| − | 1.3079 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3079 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3079 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3079 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7590 -1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7590 -1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2100 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2100 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.2100 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.2100 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7590 -0.6094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7590 -0.6094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6611 -1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6611 -1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1122 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1122 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.1122 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.1122 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6611 -0.6094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6611 -0.6094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.6611 -2.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.6611 -2.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5631 -0.6095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5631 -0.6095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0228 -0.8749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0228 -0.8749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4825 -0.6095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4825 -0.6095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4825 -0.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4825 -0.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0228 0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0228 0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.5631 -0.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.5631 -0.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.7884 -0.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.7884 -0.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.7590 -2.1708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.7590 -2.1708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.0768 0.2645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.0768 0.2645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.0228 0.7175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.0228 0.7175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1323 -0.7359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1323 -0.7359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.6166 -1.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.6166 -1.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.8741 -1.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.8741 -1.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.1577 -1.1201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.1577 -1.1201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.6783 -0.5993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.6783 -0.5993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.3279 -0.9422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.3279 -0.9422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8461 -1.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8461 -1.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.4204 -1.4557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.4204 -1.4557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.4487 -1.8420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.4487 -1.8420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9036 -0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9036 -0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9036 0.2954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9036 0.2954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9036 0.8522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9036 0.8522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4534 1.1122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4534 1.1122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3948 1.1358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3948 1.1358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8399 0.8789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8399 0.8789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3786 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3786 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.3786 1.8438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.3786 1.8438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9449 2.1708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9449 2.1708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5112 1.8438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5112 1.8438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5112 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5112 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9449 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9449 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.0768 2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.0768 2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 9 12 1 0 0 0 0 | + | 9 12 1 0 0 0 0 |
| − | 12 13 2 0 0 0 0 | + | 12 13 2 0 0 0 0 |
| − | 13 14 1 0 0 0 0 | + | 13 14 1 0 0 0 0 |
| − | 14 15 2 0 0 0 0 | + | 14 15 2 0 0 0 0 |
| − | 15 16 1 0 0 0 0 | + | 15 16 1 0 0 0 0 |
| − | 16 17 2 0 0 0 0 | + | 16 17 2 0 0 0 0 |
| − | 17 12 1 0 0 0 0 | + | 17 12 1 0 0 0 0 |
| − | 18 1 1 0 0 0 0 | + | 18 1 1 0 0 0 0 |
| − | 3 19 1 0 0 0 0 | + | 3 19 1 0 0 0 0 |
| − | 15 20 1 0 0 0 0 | + | 15 20 1 0 0 0 0 |
| − | 16 21 1 0 0 0 0 | + | 16 21 1 0 0 0 0 |
| − | 22 23 1 1 0 0 0 | + | 22 23 1 1 0 0 0 |
| − | 23 24 1 1 0 0 0 | + | 23 24 1 1 0 0 0 |
| − | 25 24 1 1 0 0 0 | + | 25 24 1 1 0 0 0 |
| − | 25 26 1 0 0 0 0 | + | 25 26 1 0 0 0 0 |
| − | 26 27 1 0 0 0 0 | + | 26 27 1 0 0 0 0 |
| − | 27 22 1 0 0 0 0 | + | 27 22 1 0 0 0 0 |
| − | 22 28 1 0 0 0 0 | + | 22 28 1 0 0 0 0 |
| − | 23 29 1 0 0 0 0 | + | 23 29 1 0 0 0 0 |
| − | 24 30 1 0 0 0 0 | + | 24 30 1 0 0 0 0 |
| − | 27 31 1 0 0 0 0 | + | 27 31 1 0 0 0 0 |
| − | 31 32 1 0 0 0 0 | + | 31 32 1 0 0 0 0 |
| − | 32 33 1 0 0 0 0 | + | 32 33 1 0 0 0 0 |
| − | 33 34 2 0 0 0 0 | + | 33 34 2 0 0 0 0 |
| − | 33 35 1 0 0 0 0 | + | 33 35 1 0 0 0 0 |
| − | 35 36 2 0 0 0 0 | + | 35 36 2 0 0 0 0 |
| − | 36 37 1 0 0 0 0 | + | 36 37 1 0 0 0 0 |
| − | 37 38 2 0 0 0 0 | + | 37 38 2 0 0 0 0 |
| − | 38 39 1 0 0 0 0 | + | 38 39 1 0 0 0 0 |
| − | 39 40 2 0 0 0 0 | + | 39 40 2 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 41 42 2 0 0 0 0 | + | 41 42 2 0 0 0 0 |
| − | 42 37 1 0 0 0 0 | + | 42 37 1 0 0 0 0 |
| − | 25 18 1 0 0 0 0 | + | 25 18 1 0 0 0 0 |
| − | 40 43 1 0 0 0 0 | + | 40 43 1 0 0 0 0 |
| − | S SKP 8 | + | S SKP 8 |
| − | ID FL3FACGS0062 | + | ID FL3FACGS0062 |
| − | KNApSAcK_ID C00004324 | + | KNApSAcK_ID C00004324 |
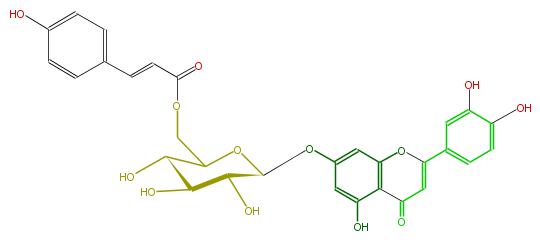
| − | NAME Luteolin 7-(6''-p-coumarylglucoside) | + | NAME Luteolin 7-(6''-p-coumarylglucoside) |
| − | CAS_RN 111188-76-2 | + | CAS_RN 111188-76-2 |
| − | FORMULA C30H26O13 | + | FORMULA C30H26O13 |
| − | EXACTMASS 594.137340918 | + | EXACTMASS 594.137340918 |
| − | AVERAGEMASS 594.51964 | + | AVERAGEMASS 594.51964 |
| − | SMILES C(C=4)(=O)c(c(OC4c(c5)cc(O)c(c5)O)1)c(O)cc(OC(O2)C(C(O)C(C2COC(C=Cc(c3)ccc(O)c3)=O)O)O)c1 | + | SMILES C(C=4)(=O)c(c(OC4c(c5)cc(O)c(c5)O)1)c(O)cc(OC(O2)C(C(O)C(C2COC(C=Cc(c3)ccc(O)c3)=O)O)O)c1 |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
43 47 0 0 0 0 0 0 0 0999 V2000
1.3079 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3079 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7590 -1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2100 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.2100 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7590 -0.6094 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6611 -1.6511 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1122 -1.3906 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.1122 -0.8698 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.6611 -0.6094 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.6611 -2.0572 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.5631 -0.6095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0228 -0.8749 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4825 -0.6095 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4825 -0.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0228 0.1868 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.5631 -0.0786 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.7884 -0.5699 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7590 -2.1708 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
5.0768 0.2645 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
4.0228 0.7175 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.1323 -0.7359 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.6166 -1.4165 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8741 -1.1278 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.1577 -1.1201 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.6783 -0.5993 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.3279 -0.9422 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8461 -1.1480 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.4204 -1.4557 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.4487 -1.8420 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9036 -0.3664 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9036 0.2954 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.9036 0.8522 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.4534 1.1122 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.3948 1.1358 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8399 0.8789 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3786 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.3786 1.8438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9449 2.1708 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5112 1.8438 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.5112 1.1899 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9449 0.8629 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.0768 2.1704 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
9 12 1 0 0 0 0
12 13 2 0 0 0 0
13 14 1 0 0 0 0
14 15 2 0 0 0 0
15 16 1 0 0 0 0
16 17 2 0 0 0 0
17 12 1 0 0 0 0
18 1 1 0 0 0 0
3 19 1 0 0 0 0
15 20 1 0 0 0 0
16 21 1 0 0 0 0
22 23 1 1 0 0 0
23 24 1 1 0 0 0
25 24 1 1 0 0 0
25 26 1 0 0 0 0
26 27 1 0 0 0 0
27 22 1 0 0 0 0
22 28 1 0 0 0 0
23 29 1 0 0 0 0
24 30 1 0 0 0 0
27 31 1 0 0 0 0
31 32 1 0 0 0 0
32 33 1 0 0 0 0
33 34 2 0 0 0 0
33 35 1 0 0 0 0
35 36 2 0 0 0 0
36 37 1 0 0 0 0
37 38 2 0 0 0 0
38 39 1 0 0 0 0
39 40 2 0 0 0 0
40 41 1 0 0 0 0
41 42 2 0 0 0 0
42 37 1 0 0 0 0
25 18 1 0 0 0 0
40 43 1 0 0 0 0
S SKP 8
ID FL3FACGS0062
KNApSAcK_ID C00004324
NAME Luteolin 7-(6''-p-coumarylglucoside)
CAS_RN 111188-76-2
FORMULA C30H26O13
EXACTMASS 594.137340918
AVERAGEMASS 594.51964
SMILES C(C=4)(=O)c(c(OC4c(c5)cc(O)c(c5)O)1)c(O)cc(OC(O2)C(C(O)C(C2COC(C=Cc(c3)ccc(O)c3)=O)O)O)c1
M END