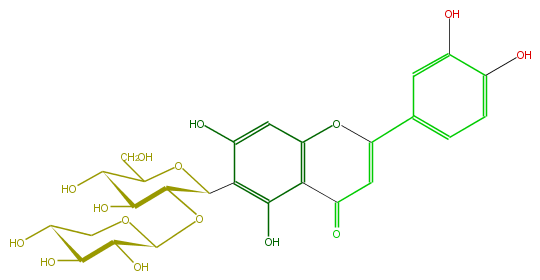
Mol:FL3FACCS0016
From Metabolomics.JP
(Difference between revisions)
| Line 1: | Line 1: | ||
| − | + | ||
| − | + | ||
| − | Copyright: ARM project http://www.metabolome.jp/ | + | Copyright: ARM project http://www.metabolome.jp/ |
| − | 41 45 0 0 0 0 0 0 0 0999 V2000 | + | 41 45 0 0 0 0 0 0 0 0999 V2000 |
| − | -0.7537 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7537 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.7537 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.7537 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0450 -1.2984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0450 -1.2984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6637 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6637 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 0.6637 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 0.6637 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0450 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0450 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3724 -1.2984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3724 -1.2984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0811 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0811 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.0811 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.0811 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3724 0.3383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3724 0.3383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1.3724 -1.9364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 1.3724 -1.9364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -0.0450 -2.1164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -0.0450 -2.1164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9472 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9472 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6941 0.0384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6941 0.0384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4409 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4409 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 4.4409 1.3319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 4.4409 1.3319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6941 1.7631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6941 1.7631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 2.9472 1.3319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | 2.9472 1.3319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 5.1872 1.7629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 5.1872 1.7629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4911 -0.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4911 -0.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.8342 -1.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.8342 -1.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.1772 -1.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.1772 -1.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.2839 -1.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.2839 -1.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.9145 -0.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.9145 -0.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.6240 -0.8746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.6240 -0.8746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.1004 -1.0162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.1004 -1.0162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.4344 -1.4001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.4344 -1.4001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4622 0.3382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4622 0.3382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -1.4707 -1.6371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -1.4707 -1.6371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 3.6941 2.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | 3.6941 2.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5814 -1.7839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5814 -1.7839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.9245 -2.5196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.9245 -2.5196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.2675 -2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.2675 -2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.3742 -2.2305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.3742 -2.2305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.0048 -1.6525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.0048 -1.6525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.7143 -1.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.7143 -1.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -5.1872 -2.1337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -5.1872 -2.1337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.5443 -2.5196 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.5443 -2.5196 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -2.7680 -2.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -2.7680 -2.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -3.1424 -0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 | + | -3.1424 -0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | -4.3632 0.0078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 | + | -4.3632 0.0078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0 |
| − | 1 2 1 0 0 0 0 | + | 1 2 1 0 0 0 0 |
| − | 2 3 2 0 0 0 0 | + | 2 3 2 0 0 0 0 |
| − | 3 4 1 0 0 0 0 | + | 3 4 1 0 0 0 0 |
| − | 4 5 2 0 0 0 0 | + | 4 5 2 0 0 0 0 |
| − | 5 6 1 0 0 0 0 | + | 5 6 1 0 0 0 0 |
| − | 6 1 2 0 0 0 0 | + | 6 1 2 0 0 0 0 |
| − | 4 7 1 0 0 0 0 | + | 4 7 1 0 0 0 0 |
| − | 7 8 1 0 0 0 0 | + | 7 8 1 0 0 0 0 |
| − | 8 9 2 0 0 0 0 | + | 8 9 2 0 0 0 0 |
| − | 9 10 1 0 0 0 0 | + | 9 10 1 0 0 0 0 |
| − | 10 5 1 0 0 0 0 | + | 10 5 1 0 0 0 0 |
| − | 7 11 2 0 0 0 0 | + | 7 11 2 0 0 0 0 |
| − | 3 12 1 0 0 0 0 | + | 3 12 1 0 0 0 0 |
| − | 13 14 2 0 0 0 0 | + | 13 14 2 0 0 0 0 |
| − | 14 15 1 0 0 0 0 | + | 14 15 1 0 0 0 0 |
| − | 15 16 2 0 0 0 0 | + | 15 16 2 0 0 0 0 |
| − | 16 17 1 0 0 0 0 | + | 16 17 1 0 0 0 0 |
| − | 17 18 2 0 0 0 0 | + | 17 18 2 0 0 0 0 |
| − | 18 13 1 0 0 0 0 | + | 18 13 1 0 0 0 0 |
| − | 9 13 1 0 0 0 0 | + | 9 13 1 0 0 0 0 |
| − | 16 19 1 0 0 0 0 | + | 16 19 1 0 0 0 0 |
| − | 20 21 1 1 0 0 0 | + | 20 21 1 1 0 0 0 |
| − | 21 22 1 1 0 0 0 | + | 21 22 1 1 0 0 0 |
| − | 23 22 1 1 0 0 0 | + | 23 22 1 1 0 0 0 |
| − | 23 24 1 0 0 0 0 | + | 23 24 1 0 0 0 0 |
| − | 24 25 1 0 0 0 0 | + | 24 25 1 0 0 0 0 |
| − | 25 20 1 0 0 0 0 | + | 25 20 1 0 0 0 0 |
| − | 20 26 1 0 0 0 0 | + | 20 26 1 0 0 0 0 |
| − | 21 27 1 0 0 0 0 | + | 21 27 1 0 0 0 0 |
| − | 2 23 1 0 0 0 0 | + | 2 23 1 0 0 0 0 |
| − | 1 28 1 0 0 0 0 | + | 1 28 1 0 0 0 0 |
| − | 22 29 1 0 0 0 0 | + | 22 29 1 0 0 0 0 |
| − | 17 30 1 0 0 0 0 | + | 17 30 1 0 0 0 0 |
| − | 31 32 1 1 0 0 0 | + | 31 32 1 1 0 0 0 |
| − | 32 33 1 1 0 0 0 | + | 32 33 1 1 0 0 0 |
| − | 34 33 1 1 0 0 0 | + | 34 33 1 1 0 0 0 |
| − | 34 35 1 0 0 0 0 | + | 34 35 1 0 0 0 0 |
| − | 35 36 1 0 0 0 0 | + | 35 36 1 0 0 0 0 |
| − | 36 31 1 0 0 0 0 | + | 36 31 1 0 0 0 0 |
| − | 31 37 1 0 0 0 0 | + | 31 37 1 0 0 0 0 |
| − | 32 38 1 0 0 0 0 | + | 32 38 1 0 0 0 0 |
| − | 33 39 1 0 0 0 0 | + | 33 39 1 0 0 0 0 |
| − | 34 29 1 0 0 0 0 | + | 34 29 1 0 0 0 0 |
| − | 40 41 1 0 0 0 0 | + | 40 41 1 0 0 0 0 |
| − | 25 40 1 0 0 0 0 | + | 25 40 1 0 0 0 0 |
| − | M STY 1 1 SUP | + | M STY 1 1 SUP |
| − | M SLB 1 1 1 | + | M SLB 1 1 1 |
| − | M SAL 1 2 40 41 | + | M SAL 1 2 40 41 |
| − | M SBL 1 1 45 | + | M SBL 1 1 45 |
| − | M SMT 1 ^ CH2OH | + | M SMT 1 ^ CH2OH |
| − | M SBV 1 45 0.5184 -0.5184 | + | M SBV 1 45 0.5184 -0.5184 |
| − | S SKP 5 | + | S SKP 5 |
| − | ID FL3FACCS0016 | + | ID FL3FACCS0016 |
| − | FORMULA C26H28O15 | + | FORMULA C26H28O15 |
| − | EXACTMASS 580.1428202259999 | + | EXACTMASS 580.1428202259999 |
| − | AVERAGEMASS 580.49152 | + | AVERAGEMASS 580.49152 |
| − | SMILES c(c3O)(c(cc(O4)c(C(C=C(c(c5)ccc(c5O)O)4)=O)3)O)C(C1OC(O2)C(C(O)C(O)C2)O)OC(CO)C(O)C1O | + | SMILES c(c3O)(c(cc(O4)c(C(C=C(c(c5)ccc(c5O)O)4)=O)3)O)C(C1OC(O2)C(C(O)C(O)C2)O)OC(CO)C(O)C1O |
M END | M END | ||
| − | |||
Latest revision as of 09:00, 14 March 2009
Copyright: ARM project http://www.metabolome.jp/
41 45 0 0 0 0 0 0 0 0999 V2000
-0.7537 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.7537 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0450 -1.2984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6637 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.6637 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.0450 0.3383 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3724 -1.2984 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0811 -0.8892 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.0811 -0.0709 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.3724 0.3383 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.3724 -1.9364 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-0.0450 -2.1164 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
2.9472 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6941 0.0384 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4409 0.4695 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.4409 1.3319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.6941 1.7631 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.9472 1.3319 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.1872 1.7629 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4911 -0.6644 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.8342 -1.4001 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.1772 -1.0061 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.2839 -1.1112 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.9145 -0.5330 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.6240 -0.8746 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.1004 -1.0162 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.4344 -1.4001 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4622 0.3382 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-1.4707 -1.6371 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
3.6941 2.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5814 -1.7839 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.9245 -2.5196 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.2675 -2.1254 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.3742 -2.2305 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-3.0048 -1.6525 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.7143 -1.9941 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-5.1872 -2.1337 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-4.5443 -2.5196 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-2.7680 -2.6250 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
-3.1424 -0.3562 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-4.3632 0.0078 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
2 3 2 0 0 0 0
3 4 1 0 0 0 0
4 5 2 0 0 0 0
5 6 1 0 0 0 0
6 1 2 0 0 0 0
4 7 1 0 0 0 0
7 8 1 0 0 0 0
8 9 2 0 0 0 0
9 10 1 0 0 0 0
10 5 1 0 0 0 0
7 11 2 0 0 0 0
3 12 1 0 0 0 0
13 14 2 0 0 0 0
14 15 1 0 0 0 0
15 16 2 0 0 0 0
16 17 1 0 0 0 0
17 18 2 0 0 0 0
18 13 1 0 0 0 0
9 13 1 0 0 0 0
16 19 1 0 0 0 0
20 21 1 1 0 0 0
21 22 1 1 0 0 0
23 22 1 1 0 0 0
23 24 1 0 0 0 0
24 25 1 0 0 0 0
25 20 1 0 0 0 0
20 26 1 0 0 0 0
21 27 1 0 0 0 0
2 23 1 0 0 0 0
1 28 1 0 0 0 0
22 29 1 0 0 0 0
17 30 1 0 0 0 0
31 32 1 1 0 0 0
32 33 1 1 0 0 0
34 33 1 1 0 0 0
34 35 1 0 0 0 0
35 36 1 0 0 0 0
36 31 1 0 0 0 0
31 37 1 0 0 0 0
32 38 1 0 0 0 0
33 39 1 0 0 0 0
34 29 1 0 0 0 0
40 41 1 0 0 0 0
25 40 1 0 0 0 0
M STY 1 1 SUP
M SLB 1 1 1
M SAL 1 2 40 41
M SBL 1 1 45
M SMT 1 ^ CH2OH
M SBV 1 45 0.5184 -0.5184
S SKP 5
ID FL3FACCS0016
FORMULA C26H28O15
EXACTMASS 580.1428202259999
AVERAGEMASS 580.49152
SMILES c(c3O)(c(cc(O4)c(C(C=C(c(c5)ccc(c5O)O)4)=O)3)O)C(C1OC(O2)C(C(O)C(O)C2)O)OC(CO)C(O)C1O
M END